pacman::p_load(sf,tidyverse)Hands-On Exercise 1
Overview
Learn how to handle geospatial data in R by using sf package.
Learn how to plot functional and truthful choropleth maps by using tmap package
1 Handle Geospatial Data using sf package
1.1 Data Acquisition
Datasets are acquired from the following areas:
Master Plan 2014 Subzone Boundary (Web) from data.gov.sg
Pre-Schools Location from data.gov.sg
Cycling Path from LTADataMall
Singapore Airbnb listing data, 19 July 2021 from Inside Airbnb
1.2 Getting Started
1.2.1 Import libraries
Two r packages need to be loaded - ‘sf’ and ‘tidyverse’
1.2.2 Import Geospatial Data using st_read() of sf package
1.2.2.1 Import polygon feature in shape file format using st_read()
When the input geospatial data is in shapefile format, two arguments will be used, namely: dsn to define the data path and layer to provide the shapefile name. Also note that no extension such as .shp, .dbf, .prj and .shx are needed.
mpsz = st_read(dsn = "Hands-On_Ex1/data/geospatial",
layer = "MP14_SUBZONE_WEB_PL") # The dsn path is with reference to the .qmd document created to house this pageReading layer `MP14_SUBZONE_WEB_PL' from data source
`C:\Cabbie-UK\ISSS624\Hands-On_Ex\Hands-On_Ex1\data\geospatial'
using driver `ESRI Shapefile'
Simple feature collection with 323 features and 15 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2667.538 ymin: 15748.72 xmax: 56396.44 ymax: 50256.33
Projected CRS: SVY21From the results above, to check that we are using “Projected CRS” for the coordinates to be reflected in a 2-D space so that we can measure distance accurately.
1.2.2.2 Import polyline feature data in shapefile format
cyclingpath = st_read(dsn = "Hands-On_Ex1/data/geospatial",
layer = "CyclingPath")Reading layer `CyclingPath' from data source
`C:\Cabbie-UK\ISSS624\Hands-On_Ex\Hands-On_Ex1\data\geospatial'
using driver `ESRI Shapefile'
Simple feature collection with 1625 features and 2 fields
Geometry type: LINESTRING
Dimension: XY
Bounding box: xmin: 12711.19 ymin: 28711.33 xmax: 42626.09 ymax: 48948.15
Projected CRS: SVY211.2.2.3 Import GIS data in kml format
preschool = st_read("Hands-On_Ex1/data/geospatial/pre-schools-location-kml.kml")Reading layer `PRESCHOOLS_LOCATION' from data source
`C:\Cabbie-UK\ISSS624\Hands-On_Ex\Hands-On_Ex1\data\geospatial\pre-schools-location-kml.kml'
using driver `KML'
Simple feature collection with 1359 features and 2 fields
Geometry type: POINT
Dimension: XYZ
Bounding box: xmin: 103.6824 ymin: 1.248403 xmax: 103.9897 ymax: 1.462134
z_range: zmin: 0 zmax: 0
Geodetic CRS: WGS 841.3 Check the content of a simple feature dataframe
Different ways to retrieve content information of a simple feature data frame
1.3.1 Use st_geometry()
To display basic information of the feature class such as type of geometry, the geographic extent of the features and the coordinate system of the data
st_geometry(mpsz)Geometry set for 323 features
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2667.538 ymin: 15748.72 xmax: 56396.44 ymax: 50256.33
Projected CRS: SVY21
First 5 geometries:MULTIPOLYGON (((31495.56 30140.01, 31980.96 296...MULTIPOLYGON (((29092.28 30021.89, 29119.64 300...MULTIPOLYGON (((29932.33 29879.12, 29947.32 298...MULTIPOLYGON (((27131.28 30059.73, 27088.33 297...MULTIPOLYGON (((26451.03 30396.46, 26440.47 303...1.3.2 Use glimpse() of dplyr
To learn more about the associated attribute information in the data frame in addition to the basic feature information.
glimpse(mpsz)Rows: 323
Columns: 16
$ OBJECTID <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, …
$ SUBZONE_NO <int> 1, 1, 3, 8, 3, 7, 9, 2, 13, 7, 12, 6, 1, 5, 1, 1, 3, 2, 2, …
$ SUBZONE_N <chr> "MARINA SOUTH", "PEARL'S HILL", "BOAT QUAY", "HENDERSON HIL…
$ SUBZONE_C <chr> "MSSZ01", "OTSZ01", "SRSZ03", "BMSZ08", "BMSZ03", "BMSZ07",…
$ CA_IND <chr> "Y", "Y", "Y", "N", "N", "N", "N", "Y", "N", "N", "N", "N",…
$ PLN_AREA_N <chr> "MARINA SOUTH", "OUTRAM", "SINGAPORE RIVER", "BUKIT MERAH",…
$ PLN_AREA_C <chr> "MS", "OT", "SR", "BM", "BM", "BM", "BM", "SR", "QT", "QT",…
$ REGION_N <chr> "CENTRAL REGION", "CENTRAL REGION", "CENTRAL REGION", "CENT…
$ REGION_C <chr> "CR", "CR", "CR", "CR", "CR", "CR", "CR", "CR", "CR", "CR",…
$ INC_CRC <chr> "5ED7EB253F99252E", "8C7149B9EB32EEFC", "C35FEFF02B13E0E5",…
$ FMEL_UPD_D <date> 2014-12-05, 2014-12-05, 2014-12-05, 2014-12-05, 2014-12-05…
$ X_ADDR <dbl> 31595.84, 28679.06, 29654.96, 26782.83, 26201.96, 25358.82,…
$ Y_ADDR <dbl> 29220.19, 29782.05, 29974.66, 29933.77, 30005.70, 29991.38,…
$ SHAPE_Leng <dbl> 5267.381, 3506.107, 1740.926, 3313.625, 2825.594, 4428.913,…
$ SHAPE_Area <dbl> 1630379.27, 559816.25, 160807.50, 595428.89, 387429.44, 103…
$ geometry <MULTIPOLYGON [m]> MULTIPOLYGON (((31495.56 30..., MULTIPOLYGON (…1.3.3 Use head()
To reveal complete information of a feature object
head(mpsz, n=5) Simple feature collection with 5 features and 15 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 25867.68 ymin: 28369.47 xmax: 32362.39 ymax: 30435.54
Projected CRS: SVY21
OBJECTID SUBZONE_NO SUBZONE_N SUBZONE_C CA_IND PLN_AREA_N
1 1 1 MARINA SOUTH MSSZ01 Y MARINA SOUTH
2 2 1 PEARL'S HILL OTSZ01 Y OUTRAM
3 3 3 BOAT QUAY SRSZ03 Y SINGAPORE RIVER
4 4 8 HENDERSON HILL BMSZ08 N BUKIT MERAH
5 5 3 REDHILL BMSZ03 N BUKIT MERAH
PLN_AREA_C REGION_N REGION_C INC_CRC FMEL_UPD_D X_ADDR
1 MS CENTRAL REGION CR 5ED7EB253F99252E 2014-12-05 31595.84
2 OT CENTRAL REGION CR 8C7149B9EB32EEFC 2014-12-05 28679.06
3 SR CENTRAL REGION CR C35FEFF02B13E0E5 2014-12-05 29654.96
4 BM CENTRAL REGION CR 3775D82C5DDBEFBD 2014-12-05 26782.83
5 BM CENTRAL REGION CR 85D9ABEF0A40678F 2014-12-05 26201.96
Y_ADDR SHAPE_Leng SHAPE_Area geometry
1 29220.19 5267.381 1630379.3 MULTIPOLYGON (((31495.56 30...
2 29782.05 3506.107 559816.2 MULTIPOLYGON (((29092.28 30...
3 29974.66 1740.926 160807.5 MULTIPOLYGON (((29932.33 29...
4 29933.77 3313.625 595428.9 MULTIPOLYGON (((27131.28 30...
5 30005.70 2825.594 387429.4 MULTIPOLYGON (((26451.03 30...1.4 Plot the Geospatial Data
To see some of the geospatial features in the dataframe, we use plot()
plot(mpsz)Warning: plotting the first 9 out of 15 attributes; use max.plot = 15 to plot
all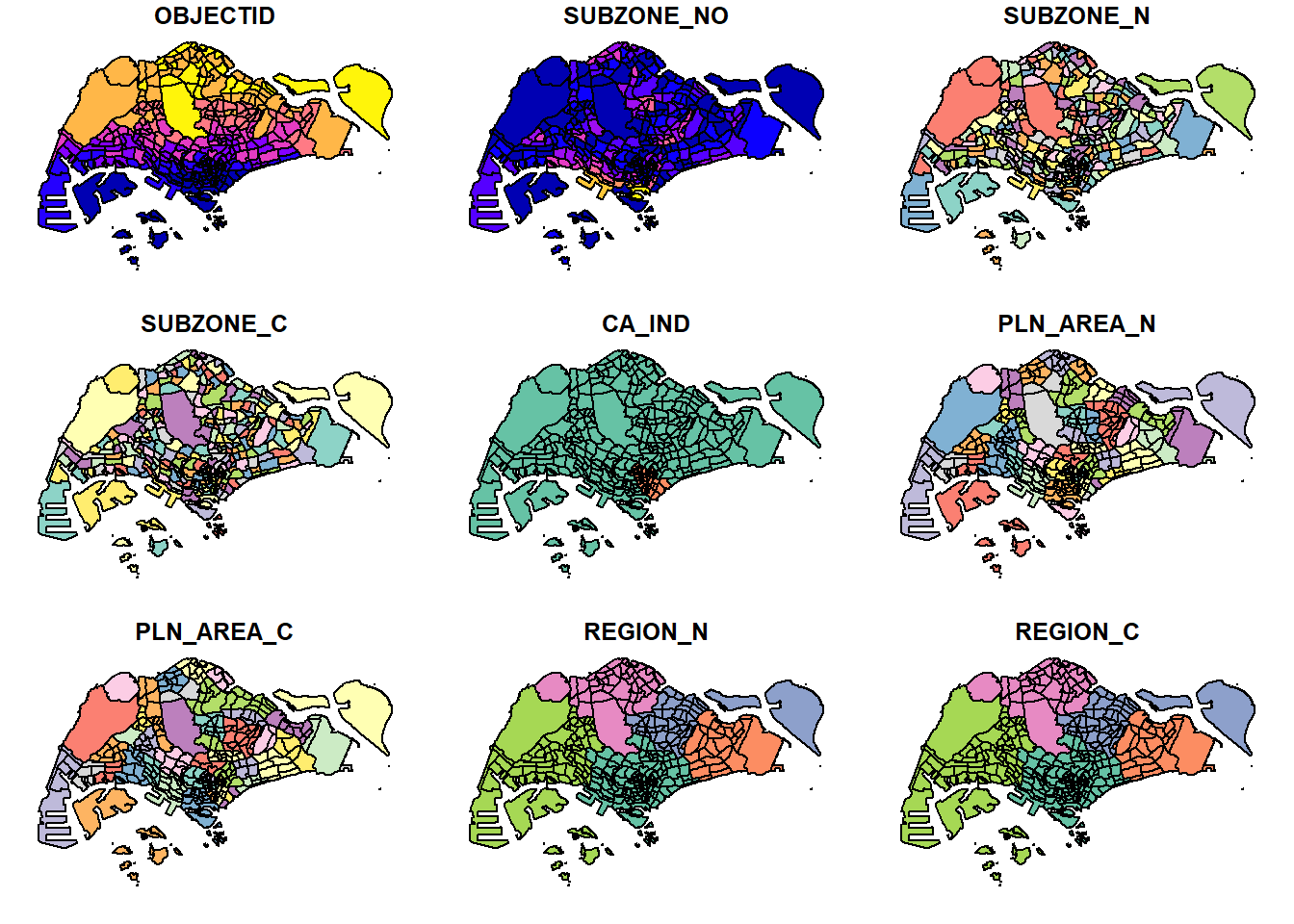
The default plot of an sf object is a multi-plot of all attributes, up to a reasonable maximum as shown above. We can, however, choose to plot only the geometry by using the code chunk below.
plot(st_geometry(mpsz))
Alternatively, we can also choose the plot the sf object by using a specific attribute as shown in the code chunk below.
plot(mpsz["REGION_C"])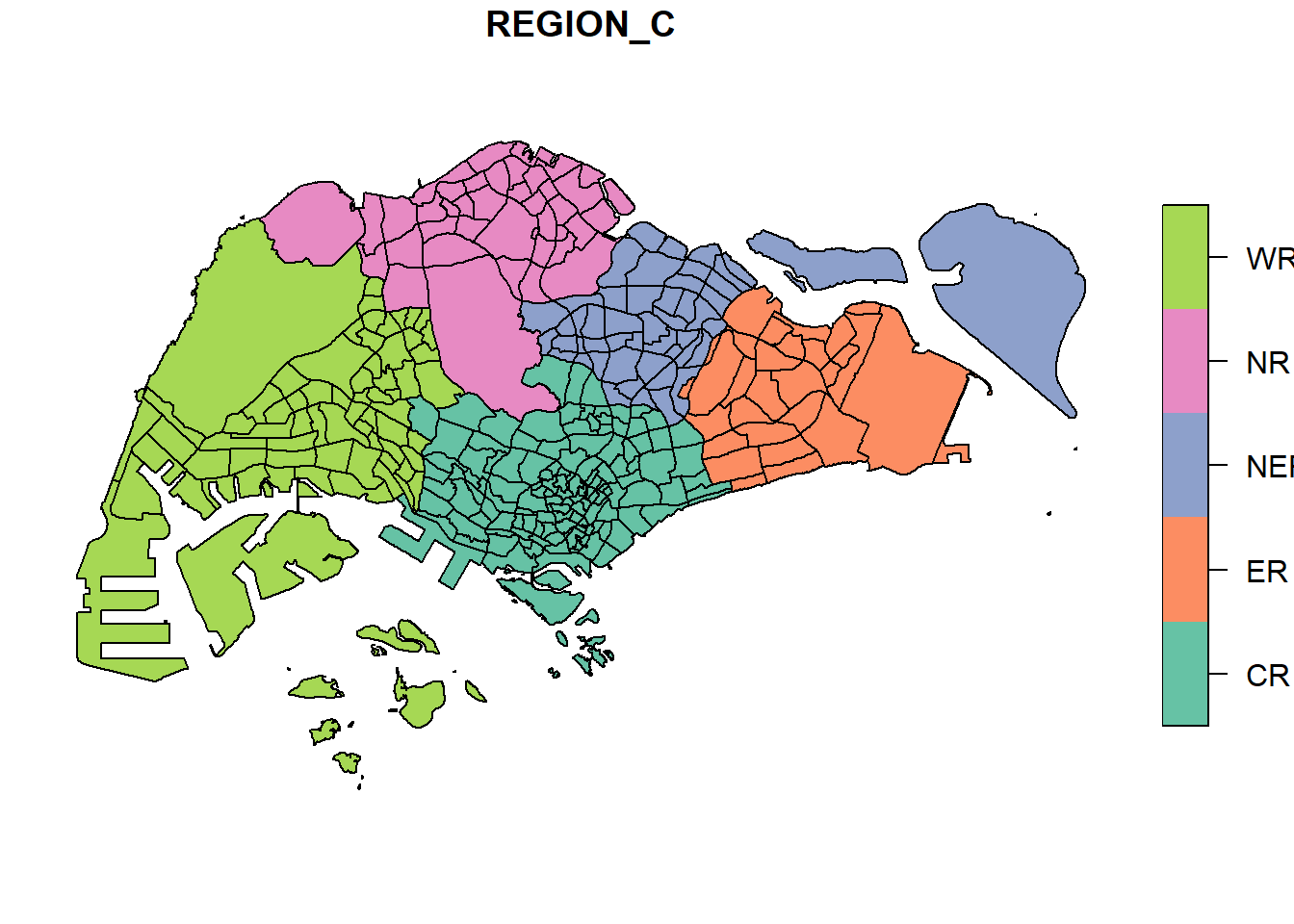
Note: plot() is mean for plotting the geospatial object for quick look. For high cartographic quality plot, other R package such as tmap should be used.
1.5 Map Projection
In a map projection, coordinates, often expressed as latitude and longitude, of locations from the surface of the globe are transformed to coordinates on a plane. Projection is a necessary step in creating a two-dimensional map and is one of the essential elements of cartography. (source: Map projection - Wikipedia)
In order to perform geoprocessing using two geospatial data, we need to ensure that both geospatial data are projected using similar coordinate system. The process to project a simple feature data frame from one coordinate system to another coordinate system is called projection transformation.
1.5.1 Assign EPSG code to a simple feature data frame
One common issue that can happen during importing geospatial data into R is that the coordinate system of the source data was either missing (such as due to missing .proj for ESRI shapefile) or wrongly assigned during the importing process.
This is an example the coordinate system of mpsz simple feature data frame by using st_crs() of sf package as shown in the code chunk below.
st_crs(mpsz)Coordinate Reference System:
User input: SVY21
wkt:
PROJCRS["SVY21",
BASEGEOGCRS["SVY21[WGS84]",
DATUM["World Geodetic System 1984",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]],
ID["EPSG",6326]],
PRIMEM["Greenwich",0,
ANGLEUNIT["Degree",0.0174532925199433]]],
CONVERSION["unnamed",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",1.36666666666667,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",103.833333333333,
ANGLEUNIT["Degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",1,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",28001.642,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",38744.572,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["(E)",east,
ORDER[1],
LENGTHUNIT["metre",1,
ID["EPSG",9001]]],
AXIS["(N)",north,
ORDER[2],
LENGTHUNIT["metre",1,
ID["EPSG",9001]]]]Although mpsz data frame is projected in svy21 but when we read until the end of the print, it indicates that the EPSG is 9001. This is a wrong EPSG code because the correct EPSG code for svy21 should be 3414.
In order to assign the correct EPSG code to mpsz data frame, st_set_crs() of sf package is used as shown in the code chunk below.
mpsz3414 <- st_set_crs(mpsz, 3414)Warning: st_crs<- : replacing crs does not reproject data; use st_transform for
thatNow, let us check the CSR again by using the code chunk below.
st_crs(mpsz3414)Coordinate Reference System:
User input: EPSG:3414
wkt:
PROJCRS["SVY21 / Singapore TM",
BASEGEOGCRS["SVY21",
DATUM["SVY21",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4757]],
CONVERSION["Singapore Transverse Mercator",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",1.36666666666667,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",103.833333333333,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",1,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",28001.642,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",38744.572,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["northing (N)",north,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["easting (E)",east,
ORDER[2],
LENGTHUNIT["metre",1]],
USAGE[
SCOPE["Cadastre, engineering survey, topographic mapping."],
AREA["Singapore - onshore and offshore."],
BBOX[1.13,103.59,1.47,104.07]],
ID["EPSG",3414]]1.5.2 Transform the projection of preschool from wgs84 to svy21
In geospatial analytics, it is very common for us to transform the original data from geographic coordinate system to projected coordinate system when the analysis need to use distance or/and area measurements.
Let us take preschool simple feature data frame as an example. The print below reveals that it is in wgs84 coordinate system.
st_crs(preschool)Coordinate Reference System:
User input: WGS 84
wkt:
GEOGCRS["WGS 84",
DATUM["World Geodetic System 1984",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
CS[ellipsoidal,2],
AXIS["geodetic latitude (Lat)",north,
ORDER[1],
ANGLEUNIT["degree",0.0174532925199433]],
AXIS["geodetic longitude (Lon)",east,
ORDER[2],
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4326]]Let us perform the projection transformation by using the code chunk below with st_transform()
preschool3414 <- st_transform(preschool,
crs = 3414)Let us display the content of preschool3414 sf data frame as shown below. The coordinate system is now EPSG:3414
st_crs(preschool3414)Coordinate Reference System:
User input: EPSG:3414
wkt:
PROJCRS["SVY21 / Singapore TM",
BASEGEOGCRS["SVY21",
DATUM["SVY21",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4757]],
CONVERSION["Singapore Transverse Mercator",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",1.36666666666667,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",103.833333333333,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",1,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",28001.642,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",38744.572,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["northing (N)",north,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["easting (E)",east,
ORDER[2],
LENGTHUNIT["metre",1]],
USAGE[
SCOPE["Cadastre, engineering survey, topographic mapping."],
AREA["Singapore - onshore and offshore."],
BBOX[1.13,103.59,1.47,104.07]],
ID["EPSG",3414]]1.6 Import and convert an aspatial datafile
Aspatial data it is not a geospatial data but among the data fields, there are two fields that capture the x- and y-coordinates of the data points.
1.6.1 Import aspatial data
We will use read_csv() of readr package to import listing.csv and display part of the list as shown the code chunk below.
listings <- read_csv("Hands-On_Ex1/data/aspatial/listings.csv")Rows: 4252 Columns: 16
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (5): name, host_name, neighbourhood_group, neighbourhood, room_type
dbl (10): id, host_id, latitude, longitude, price, minimum_nights, number_o...
date (1): last_review
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.list(listings)[[1]]
# A tibble: 4,252 × 16
id name host_id host_…¹ neigh…² neigh…³ latit…⁴ longi…⁵ room_…⁶ price
<dbl> <chr> <dbl> <chr> <chr> <chr> <dbl> <dbl> <chr> <dbl>
1 50646 Pleasan… 227796 Sujatha Centra… Bukit … 1.33 104. Privat… 80
2 71609 Ensuite… 367042 Belinda East R… Tampin… 1.35 104. Privat… 178
3 71896 B&B Ro… 367042 Belinda East R… Tampin… 1.35 104. Privat… 81
4 71903 Room 2-… 367042 Belinda East R… Tampin… 1.35 104. Privat… 81
5 275343 Conveni… 1439258 Joyce Centra… Bukit … 1.29 104. Privat… 52
6 275344 15 mins… 1439258 Joyce Centra… Bukit … 1.29 104. Privat… 40
7 294281 5 mins … 1521514 Elizab… Centra… Newton 1.31 104. Privat… 72
8 301247 Nice ro… 1552002 Rahul Centra… Geylang 1.32 104. Privat… 41
9 324945 20 Mins… 1439258 Joyce Centra… Bukit … 1.29 104. Privat… 49
10 330089 Accomo@… 1439258 Joyce Centra… Bukit … 1.29 104. Privat… 49
# … with 4,242 more rows, 6 more variables: minimum_nights <dbl>,
# number_of_reviews <dbl>, last_review <date>, reviews_per_month <dbl>,
# calculated_host_listings_count <dbl>, availability_365 <dbl>, and
# abbreviated variable names ¹host_name, ²neighbourhood_group,
# ³neighbourhood, ⁴latitude, ⁵longitude, ⁶room_typeThe output reveals that listing tibble data frame consists of 4252 rows and 16 columns. Two useful fields we are going to use in the next phase are latitude and longitude.
1.6.2 Create a simple feature data frame from an aspatial data frame
The code chunk below converts listing data frame into a simple feature data frame by using st_as_sf()
listings_sf <- st_as_sf(listings,
coords = c("longitude", "latitude"),
crs=4326) %>%
st_transform(crs = 3414)Things to learn from the arguments above:
coords argument requires you to provide the column name of the x-coordinates first then followed by the column name of the y-coordinates.
crs argument requires you to provide the coordinates system in epsg format. EPSG: 4326 is wgs84 Geographic Coordinate System and EPSG: 3414 is Singapore SVY21 Projected Coordinate System. You can search for other country’s epsg code by referring to epsg.io.
%>% is used to nest st_transform() to transform the newly created simple feature data frame into svy21 projected coordinates system.
Let us examine the content of this newly created simple feature data frame.
glimpse(listings_sf)Rows: 4,252
Columns: 15
$ id <dbl> 50646, 71609, 71896, 71903, 275343, 275…
$ name <chr> "Pleasant Room along Bukit Timah", "Ens…
$ host_id <dbl> 227796, 367042, 367042, 367042, 1439258…
$ host_name <chr> "Sujatha", "Belinda", "Belinda", "Belin…
$ neighbourhood_group <chr> "Central Region", "East Region", "East …
$ neighbourhood <chr> "Bukit Timah", "Tampines", "Tampines", …
$ room_type <chr> "Private room", "Private room", "Privat…
$ price <dbl> 80, 178, 81, 81, 52, 40, 72, 41, 49, 49…
$ minimum_nights <dbl> 90, 90, 90, 90, 14, 14, 90, 8, 14, 14, …
$ number_of_reviews <dbl> 18, 20, 24, 48, 20, 13, 133, 105, 14, 1…
$ last_review <date> 2014-07-08, 2019-12-28, 2014-12-10, 20…
$ reviews_per_month <dbl> 0.22, 0.28, 0.33, 0.67, 0.20, 0.16, 1.2…
$ calculated_host_listings_count <dbl> 1, 4, 4, 4, 50, 50, 7, 1, 50, 50, 50, 4…
$ availability_365 <dbl> 365, 365, 365, 365, 353, 364, 365, 90, …
$ geometry <POINT [m]> POINT (22646.02 35167.9), POINT (…1.7 Geoprocessing with sf package
sf package also offers a wide range of geoprocessing (also known as GIS analysis) functions.
1.7.1 Buffering
The scenario:
The authority is planning to upgrade the exiting cycling path. To do so, they need to acquire 5 metres of reserved land on the both sides of the current cycling path. You are tasked to determine the extend of the land need to be acquired and their total area.
The solution:
Firstly, st_buffer() of sf package is used to compute the 5-meter buffers around cycling paths
buffer_cycling <- st_buffer(cyclingpath,
dist=5, nQuadSegs = 30)This is followed by calculating the area of the buffers as shown in the code chunk below.
buffer_cycling$AREA <- st_area(buffer_cycling)Lastly, sum() of Base R will be used to derive the total land involved
sum(buffer_cycling$AREA)773143.9 [m^2]1.7.2 Point-in-polygon count
The scenario:
A pre-school service group want to find out the numbers of pre-schools in each Planning Subzone.
The solution:
The code chunk below performs two operations at one go. Firstly, identify pre-schools located inside each Planning Subzone by using st_intersects(). Next, length() of Base R is used to calculate numbers of pre-schools that fall inside each planning subzone.
mpsz3414$`PreSch Count`<- lengths(st_intersects(mpsz3414, preschool3414))To list the planning subzone with the most number of pre-school, the top_n() of dplyr package is used as shown in the code chunk below.
top_n(mpsz3414, 1, `PreSch Count`)Simple feature collection with 1 feature and 16 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 23449.05 ymin: 46001.23 xmax: 25594.22 ymax: 47996.47
Projected CRS: SVY21 / Singapore TM
OBJECTID SUBZONE_NO SUBZONE_N SUBZONE_C CA_IND PLN_AREA_N PLN_AREA_C
1 290 3 WOODLANDS EAST WDSZ03 N WOODLANDS WD
REGION_N REGION_C INC_CRC FMEL_UPD_D X_ADDR Y_ADDR
1 NORTH REGION NR C90769E43EE6B0F2 2014-12-05 24506.64 46991.63
SHAPE_Leng SHAPE_Area geometry PreSch Count
1 6603.608 2553464 MULTIPOLYGON (((24786.75 46... 37The scenario:
Calculate the density of pre-school by planning subzone
The solution:
Firstly, the code chunk below uses st_area() of sf package to derive the area of each planning subzone.
mpsz3414$Area <- mpsz3414 %>%
st_area()Next, mutate() of dplyr package is used to compute the density by using the code chunk below.
mpsz3414 <- mpsz3414 %>%
mutate(`PreSch Density` = `PreSch Count`/Area * 1000000)A peek at the resutling dataframe
head(mpsz3414)Simple feature collection with 6 features and 18 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 24468.89 ymin: 28369.47 xmax: 32362.39 ymax: 30542.74
Projected CRS: SVY21 / Singapore TM
OBJECTID SUBZONE_NO SUBZONE_N SUBZONE_C CA_IND PLN_AREA_N
1 1 1 MARINA SOUTH MSSZ01 Y MARINA SOUTH
2 2 1 PEARL'S HILL OTSZ01 Y OUTRAM
3 3 3 BOAT QUAY SRSZ03 Y SINGAPORE RIVER
4 4 8 HENDERSON HILL BMSZ08 N BUKIT MERAH
5 5 3 REDHILL BMSZ03 N BUKIT MERAH
6 6 7 ALEXANDRA HILL BMSZ07 N BUKIT MERAH
PLN_AREA_C REGION_N REGION_C INC_CRC FMEL_UPD_D X_ADDR
1 MS CENTRAL REGION CR 5ED7EB253F99252E 2014-12-05 31595.84
2 OT CENTRAL REGION CR 8C7149B9EB32EEFC 2014-12-05 28679.06
3 SR CENTRAL REGION CR C35FEFF02B13E0E5 2014-12-05 29654.96
4 BM CENTRAL REGION CR 3775D82C5DDBEFBD 2014-12-05 26782.83
5 BM CENTRAL REGION CR 85D9ABEF0A40678F 2014-12-05 26201.96
6 BM CENTRAL REGION CR 9D286521EF5E3B59 2014-12-05 25358.82
Y_ADDR SHAPE_Leng SHAPE_Area geometry PreSch Count
1 29220.19 5267.381 1630379.3 MULTIPOLYGON (((31495.56 30... 0
2 29782.05 3506.107 559816.2 MULTIPOLYGON (((29092.28 30... 5
3 29974.66 1740.926 160807.5 MULTIPOLYGON (((29932.33 29... 0
4 29933.77 3313.625 595428.9 MULTIPOLYGON (((27131.28 30... 2
5 30005.70 2825.594 387429.4 MULTIPOLYGON (((26451.03 30... 1
6 29991.38 4428.913 1030378.8 MULTIPOLYGON (((25899.7 297... 10
Area PreSch Density
1 1630379.3 [m^2] 0.000000 [1/m^2]
2 559816.2 [m^2] 8.931502 [1/m^2]
3 160807.5 [m^2] 0.000000 [1/m^2]
4 595428.9 [m^2] 3.358923 [1/m^2]
5 387429.4 [m^2] 2.581115 [1/m^2]
6 1030378.8 [m^2] 9.705169 [1/m^2]1.8 Explorotary Data Analysis (EDA)
We will use appropriate ggplot2 functions to create functional and yet truthful statistical graphs for EDA purposes.
Firstly, we will plot a histogram to reveal the distribution of PreSch Density. Conventionally, hist() of R Graphics will be used as shown in the code chunk below.
hist(mpsz3414$`PreSch Density`)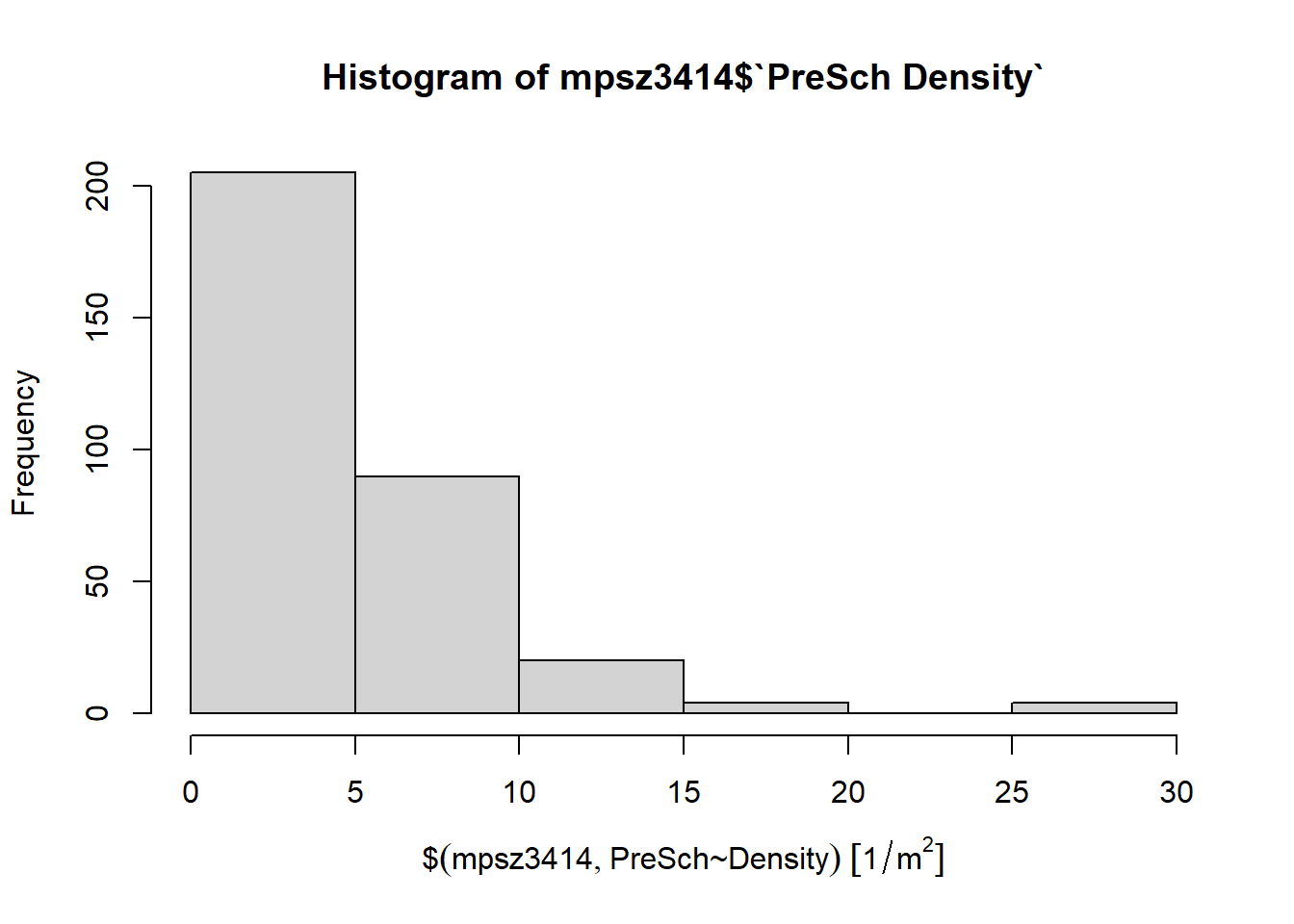
Although the syntax is very easy to use however the output is far from meeting publication quality. Furthermore, the function has limited room for further customisation.
In the code chunk below, appropriate ggplot2 functions will be used.
ggplot(data=mpsz3414,
aes(x= as.numeric(`PreSch Density`)))+
geom_histogram(bins=20,
color="black",
fill="light blue") +
labs(title = "Are pre-school even distributed in Singapore?",
subtitle= "There are many planning sub-zones with a single pre-school, on the other hand, \nthere are two planning sub-zones with at least 20 pre-schools",
x = "Pre-school density (per km sq)",
y = "Frequency")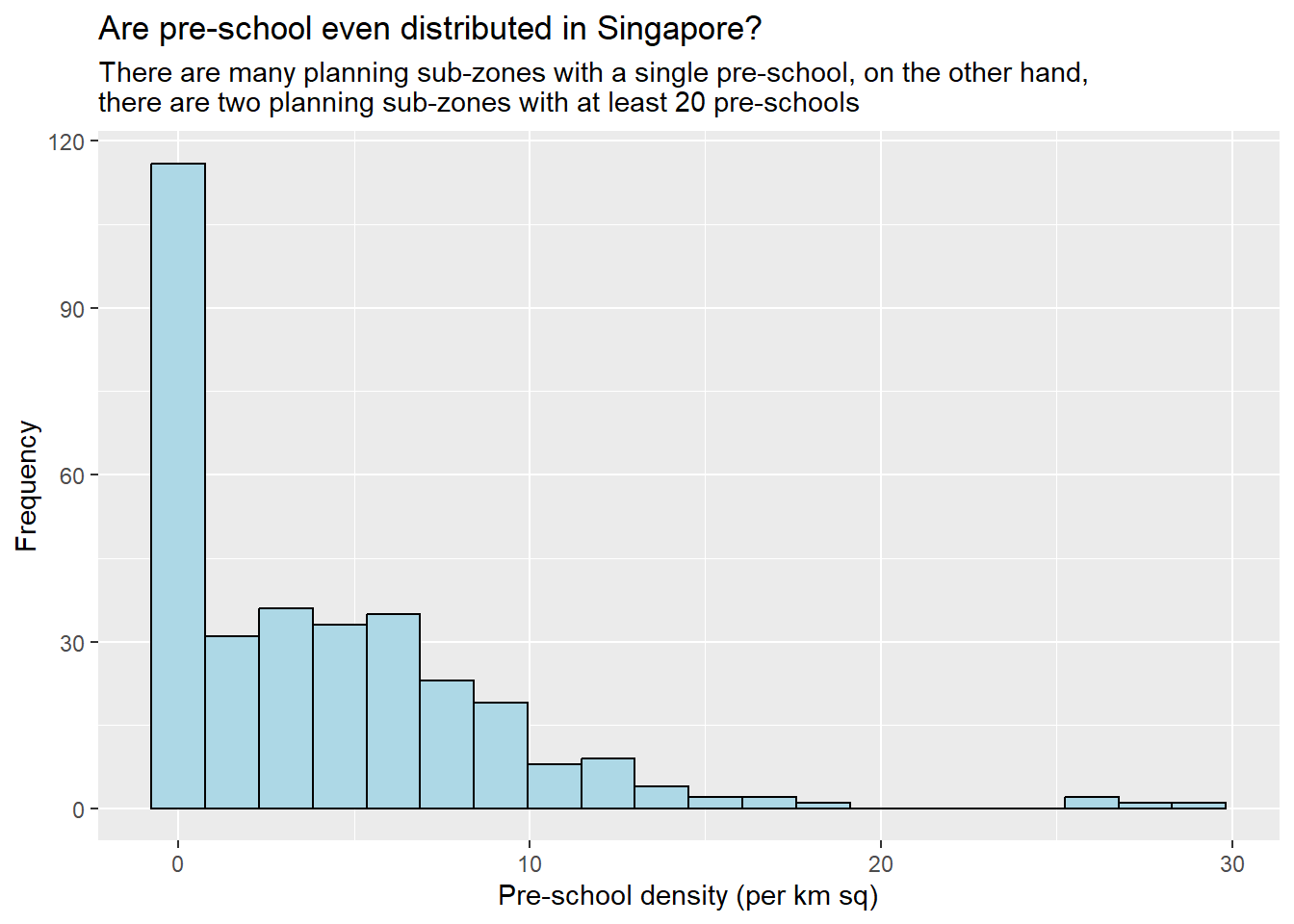
This is a scatterplot showing the relationship between Pre-school Density and Pre-school Count
ggplot(data=mpsz3414,
aes(x= as.numeric(`PreSch Density`),y=as.numeric(`PreSch Count`)))+
geom_point() +
labs(title = "Pre-School Count vs Pre-School Density",
x = "Pre-school density (per km sq)",
y = "Pre-school count")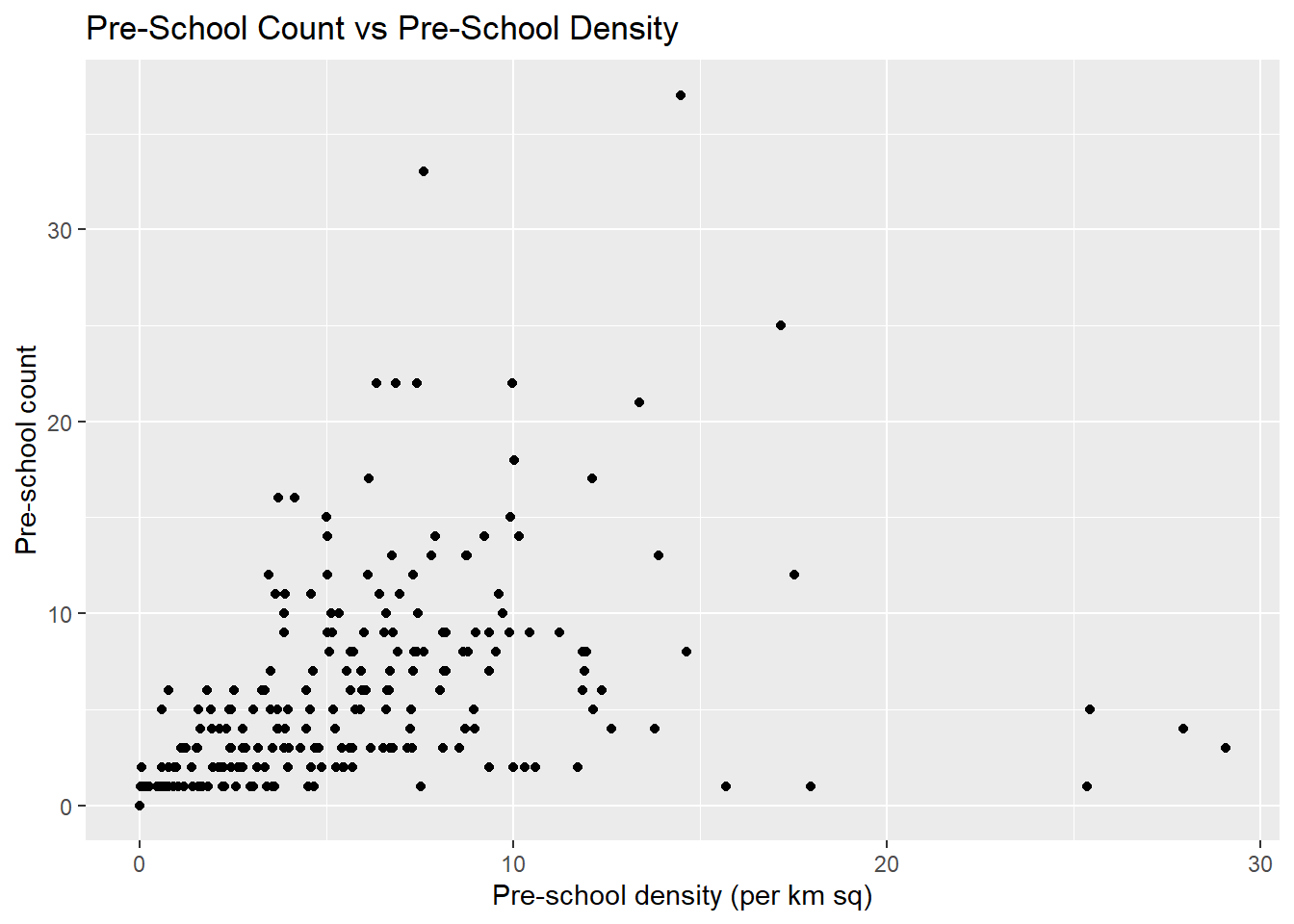
2 Choropleth Mapping with R
2.1 Import data into R
2.1.1 The data
Two data set will be used to create the choropleth map. They are:
Master Plan 2014 Subzone Boundary (Web) (i.e.
MP14_SUBZONE_WEB_PL) in ESRI shapefile format. It can be downloaded at data.gov.sg This is a geospatial data. It consists of the geographical boundary of Singapore at the planning subzone level. The data is based on URA Master Plan 2014.Singapore Residents by Planning Area / Subzone, Age Group, Sex and Type of Dwelling, June 2011-2020 in csv format (i.e.
respopagesextod2011to2020.csv). This is an aspatial data fie. It can be downloaded at Department of Statistics, Singapore Although it does not contain any coordinates values, but it’s PA and SZ fields can be used as unique identifiers to geocode toMP14_SUBZONE_WEB_PLshapefile.
2.1.2 Import Geospatial Data into R
The code chunk below uses the st_read() function of sf package to import MP14_SUBZONE_WEB_PL shapefile into R as a simple feature data frame called mpsz.
mpsz <- st_read(dsn = "Hands-On_Ex1/data/geospatial",
layer = "MP14_SUBZONE_WEB_PL")Reading layer `MP14_SUBZONE_WEB_PL' from data source
`C:\Cabbie-UK\ISSS624\Hands-On_Ex\Hands-On_Ex1\data\geospatial'
using driver `ESRI Shapefile'
Simple feature collection with 323 features and 15 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2667.538 ymin: 15748.72 xmax: 56396.44 ymax: 50256.33
Projected CRS: SVY21You can examine the content of mpsz by using the code chunk below.
mpszSimple feature collection with 323 features and 15 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 2667.538 ymin: 15748.72 xmax: 56396.44 ymax: 50256.33
Projected CRS: SVY21
First 10 features:
OBJECTID SUBZONE_NO SUBZONE_N SUBZONE_C CA_IND PLN_AREA_N
1 1 1 MARINA SOUTH MSSZ01 Y MARINA SOUTH
2 2 1 PEARL'S HILL OTSZ01 Y OUTRAM
3 3 3 BOAT QUAY SRSZ03 Y SINGAPORE RIVER
4 4 8 HENDERSON HILL BMSZ08 N BUKIT MERAH
5 5 3 REDHILL BMSZ03 N BUKIT MERAH
6 6 7 ALEXANDRA HILL BMSZ07 N BUKIT MERAH
7 7 9 BUKIT HO SWEE BMSZ09 N BUKIT MERAH
8 8 2 CLARKE QUAY SRSZ02 Y SINGAPORE RIVER
9 9 13 PASIR PANJANG 1 QTSZ13 N QUEENSTOWN
10 10 7 QUEENSWAY QTSZ07 N QUEENSTOWN
PLN_AREA_C REGION_N REGION_C INC_CRC FMEL_UPD_D X_ADDR
1 MS CENTRAL REGION CR 5ED7EB253F99252E 2014-12-05 31595.84
2 OT CENTRAL REGION CR 8C7149B9EB32EEFC 2014-12-05 28679.06
3 SR CENTRAL REGION CR C35FEFF02B13E0E5 2014-12-05 29654.96
4 BM CENTRAL REGION CR 3775D82C5DDBEFBD 2014-12-05 26782.83
5 BM CENTRAL REGION CR 85D9ABEF0A40678F 2014-12-05 26201.96
6 BM CENTRAL REGION CR 9D286521EF5E3B59 2014-12-05 25358.82
7 BM CENTRAL REGION CR 7839A8577144EFE2 2014-12-05 27680.06
8 SR CENTRAL REGION CR 48661DC0FBA09F7A 2014-12-05 29253.21
9 QT CENTRAL REGION CR 1F721290C421BFAB 2014-12-05 22077.34
10 QT CENTRAL REGION CR 3580D2AFFBEE914C 2014-12-05 24168.31
Y_ADDR SHAPE_Leng SHAPE_Area geometry
1 29220.19 5267.381 1630379.3 MULTIPOLYGON (((31495.56 30...
2 29782.05 3506.107 559816.2 MULTIPOLYGON (((29092.28 30...
3 29974.66 1740.926 160807.5 MULTIPOLYGON (((29932.33 29...
4 29933.77 3313.625 595428.9 MULTIPOLYGON (((27131.28 30...
5 30005.70 2825.594 387429.4 MULTIPOLYGON (((26451.03 30...
6 29991.38 4428.913 1030378.8 MULTIPOLYGON (((25899.7 297...
7 30230.86 3275.312 551732.0 MULTIPOLYGON (((27746.95 30...
8 30222.86 2208.619 290184.7 MULTIPOLYGON (((29351.26 29...
9 29893.78 6571.323 1084792.3 MULTIPOLYGON (((20996.49 30...
10 30104.18 3454.239 631644.3 MULTIPOLYGON (((24472.11 29...2.1.3 Import attribute data into R
We will import respopagsex2000to2018.csv file into RStudio and save the file into an R dataframe called popagsex.
popdata <- read_csv("Hands-On_Ex1/data/aspatial/respopagesextod2011to2020.csv",show_col_types = FALSE)2.1.4 Data Preparation
We need to prepare a data table that includes the variables PA, SZ, YOUNG, ECONOMY ACTIVE, AGED, TOTAL, DEPENDENCY.
YOUNG: age group 0 to 4 until age groyup 20 to 24,
ECONOMY ACTIVE: age group 25-29 until age group 60-64,
AGED: age group 65 and above,
TOTAL: all age group, and
DEPENDENCY: the ratio between young and aged against economy active group
2.1.4.1 Data Wrangling
The following data wrangling and transformation functions will be used:
pivot_wider() of tidyr package, and
mutate(), filter(), group_by() and select() of dplyr package
popdata2020 = popdata %>%
filter(Time==2020) %>%
group_by(PA, SZ, AG) %>%
summarise(`POP`=sum(`Pop`)) %>%
ungroup() %>%
pivot_wider(names_from = AG, values_from = POP) %>%
mutate(YOUNG = rowSums(.[3:6]) + rowSums(.[12])) %>%
mutate(`ECONOMIC ACTIVE`= rowSums(.[7:11]) + rowSums(.[13:15])) %>%
mutate(`AGED`=rowSums(.[16:21])) %>%
mutate(`TOTAL`=rowSums(.[3:21])) %>%
mutate(`DEPENDENCY`= (`YOUNG`+`AGED`)/`ECONOMIC ACTIVE`) %>%
select(`PA`,`SZ`,`YOUNG`,`ECONOMIC ACTIVE`,`AGED`,`TOTAL`,`DEPENDENCY`)`summarise()` has grouped output by 'PA', 'SZ'. You can override using the
`.groups` argument.2.1.4.1 Join the attribute data and geospatial data
Before we can perform the georelational join, one extra step is required to convert the values in PA and SZ fields to uppercase. This is because the values of PA and SZ fields are made up of upper- and lowercase. On the other, hand the SUBZONE_N and PLN_AREA_N are in uppercase.
popdata2020 = popdata2020 %>%
mutate_at(.vars = vars(PA, SZ),
.funs = funs(toupper)) %>%
filter(`ECONOMIC ACTIVE` > 0)Warning: `funs()` was deprecated in dplyr 0.8.0.
ℹ Please use a list of either functions or lambdas:
# Simple named list: list(mean = mean, median = median)
# Auto named with `tibble::lst()`: tibble::lst(mean, median)
# Using lambdas list(~ mean(., trim = .2), ~ median(., na.rm = TRUE))Next, left_join() of dplyr is used to join the geographical data and attribute table using planning subzone name e.g. SUBZONE_N and SZ as the common identifier.
mpsz_pop2020 = left_join(mpsz,popdata2020,
by =c("SUBZONE_N"="SZ"))Thing to learn from the code chunk above:
- left_join() of dplyr package is used with
mpszsimple feature data frame as the left data table is to ensure that the output will be a simple features data frame.
write_rds(mpsz_pop2020, "Hands-On_Ex1/data/rds/mpszpop2020.rds")2.2 Choropleth Mapping Geospatial Data Using tmap
Two ways to prepare thematic map using tmap, they are:
Plotting a thematic map quickly by using qtm().
Plotting highly customisable thematic map by using tmap elements
pacman::p_load(tmap) # load tmap package2.2.1 Quick plot using qtm()
It is concise and provides a good default visualisation in many cases. The code chunk below will draw a cartographic standard choropleth map as shown below.
tmap_mode("plot")tmap mode set to plottingqtm(mpsz_pop2020,
fill = "DEPENDENCY")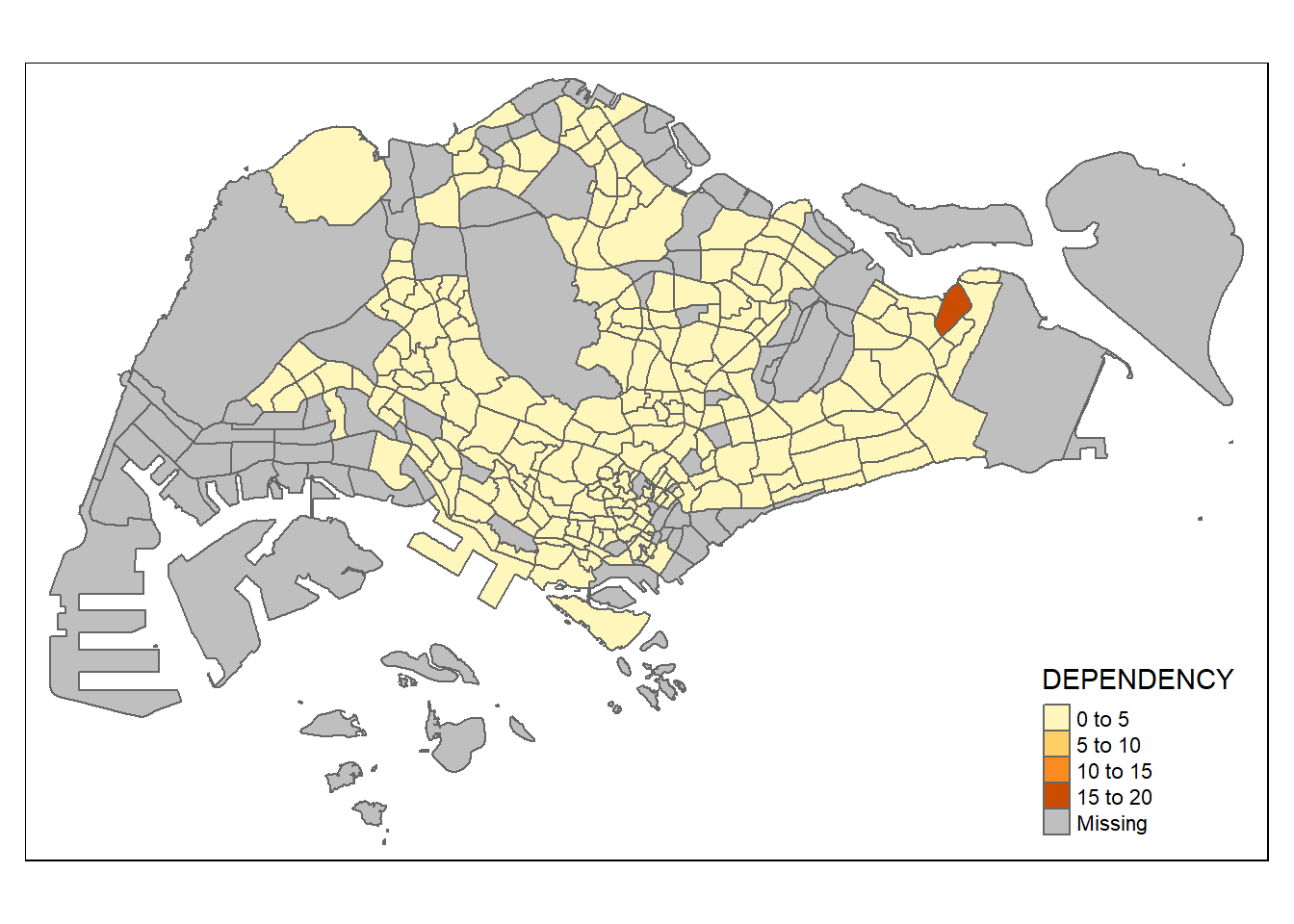
Things to note from the code chunk above:
tmap_mode() with “plot” option is used to produce a static map. For interactive mode, “view” option should be used.
fill argument is used to map the attribute (i.e. DEPENDENCY)
2.2.2 Create a choropleth map by using tmap’s elements
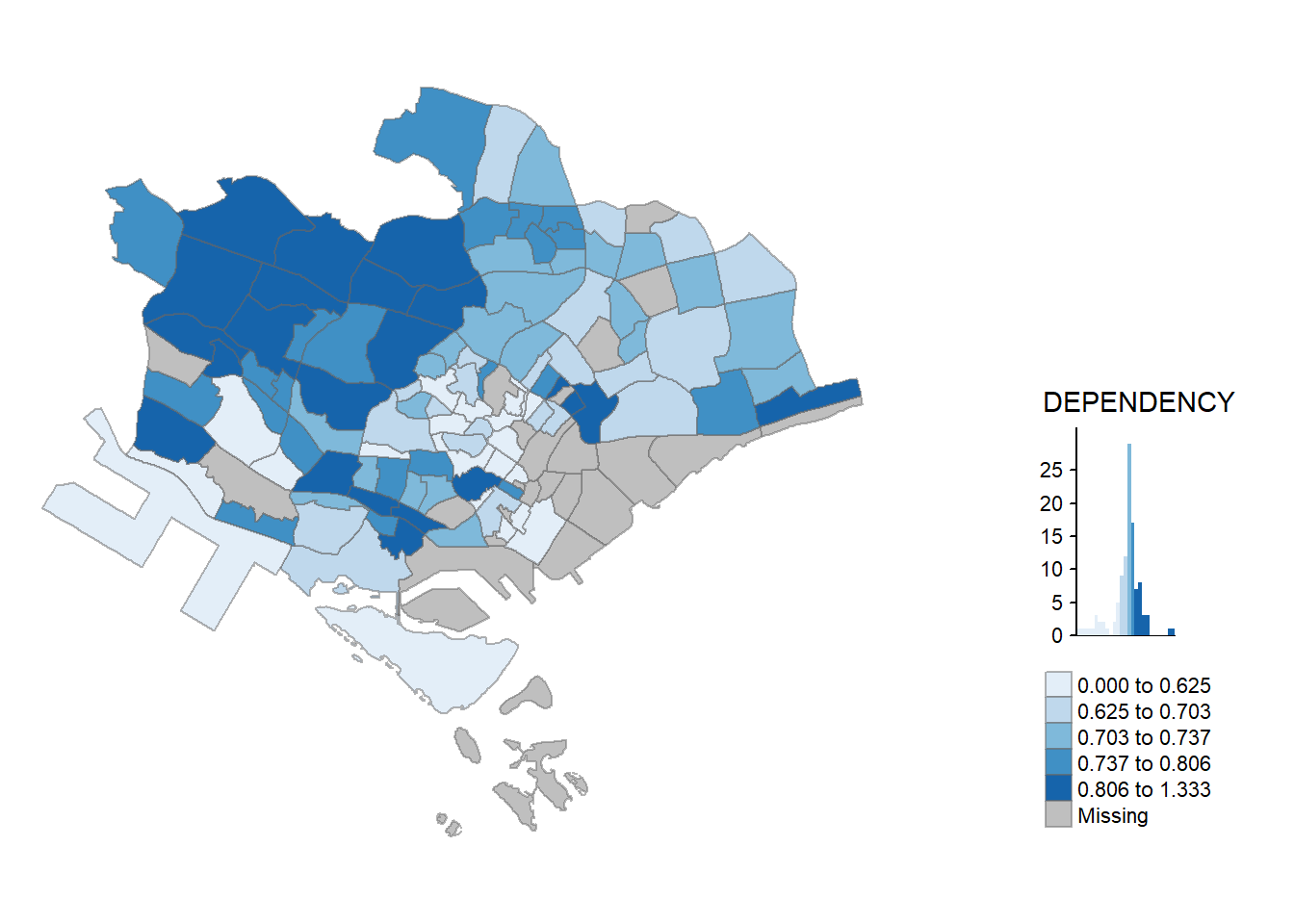
qtm() makes aesthetics of individual layers harder to control. To draw a high quality cartographic choropleth map as shown in the figure below, tmap’s drawing elements should be used.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
style = "quantile",
palette = "Blues",
title = "Dependency ratio") +
tm_layout(main.title = "Distribution of Dependency Ratio by planning subzone",
main.title.position = "center",
main.title.size = 1.2,
legend.height = 0.45,
legend.width = 0.35,
frame = TRUE) +
tm_borders(alpha = 0.5) +
tm_compass(type="8star", size = 2) +
tm_scale_bar() +
tm_grid(alpha =0.2) +
tm_credits("Source: Planning Sub-zone boundary from Urban Redevelopment Authorithy (URA)\n and Population data from Department of Statistics DOS",
position = c("left", "bottom"))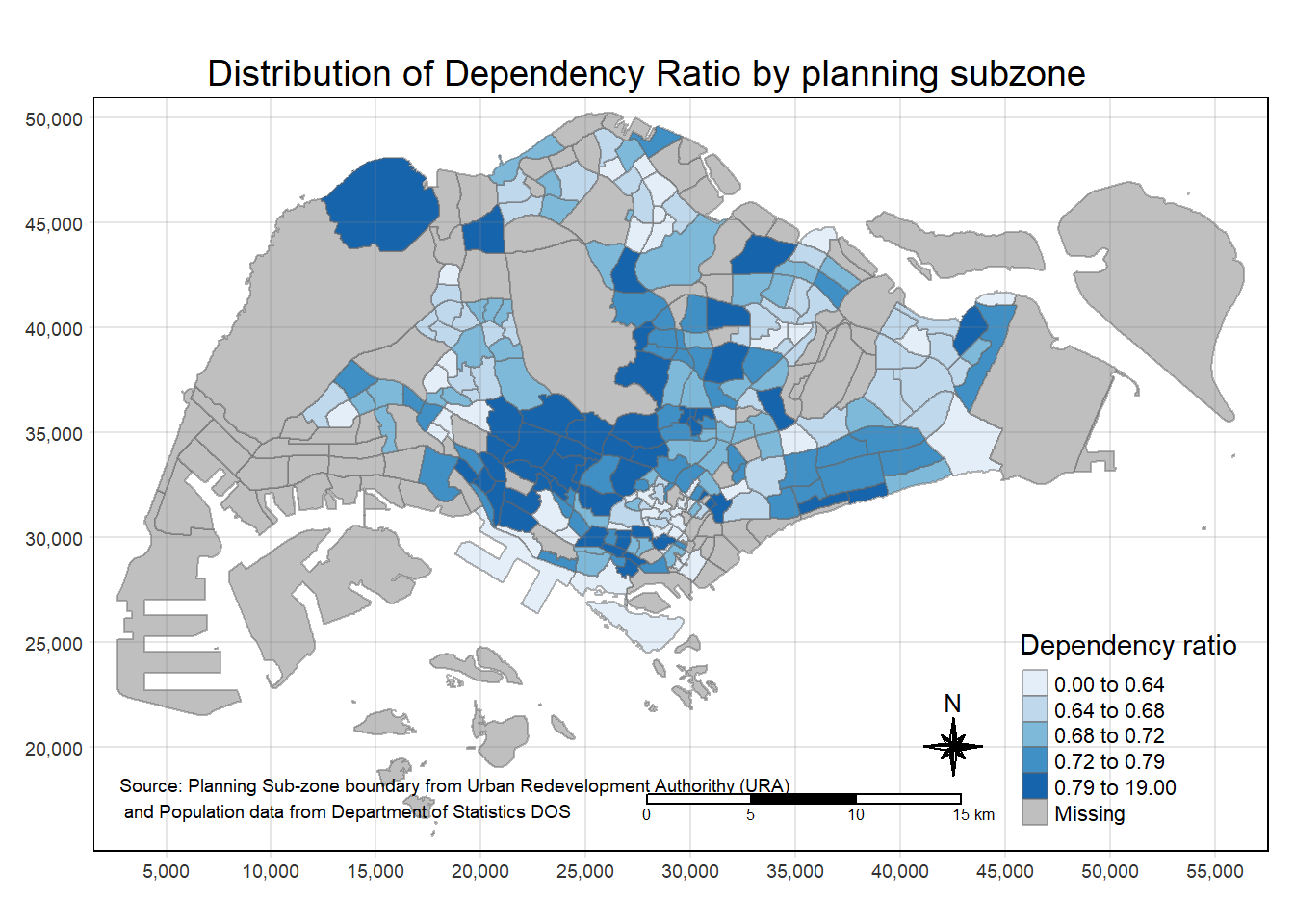
2.2.2.1 Draw a base map
The basic building block of tmap is tm_shape() followed by one or more layer elemments such as tm_fill() and tm_polygons().
In the code chunk below, tm_shape() is used to define the input data (i.e mpsz_pop2020) and tm_polygons() is used to draw the planning subzone polygons
tm_shape(mpsz_pop2020) +
tm_polygons()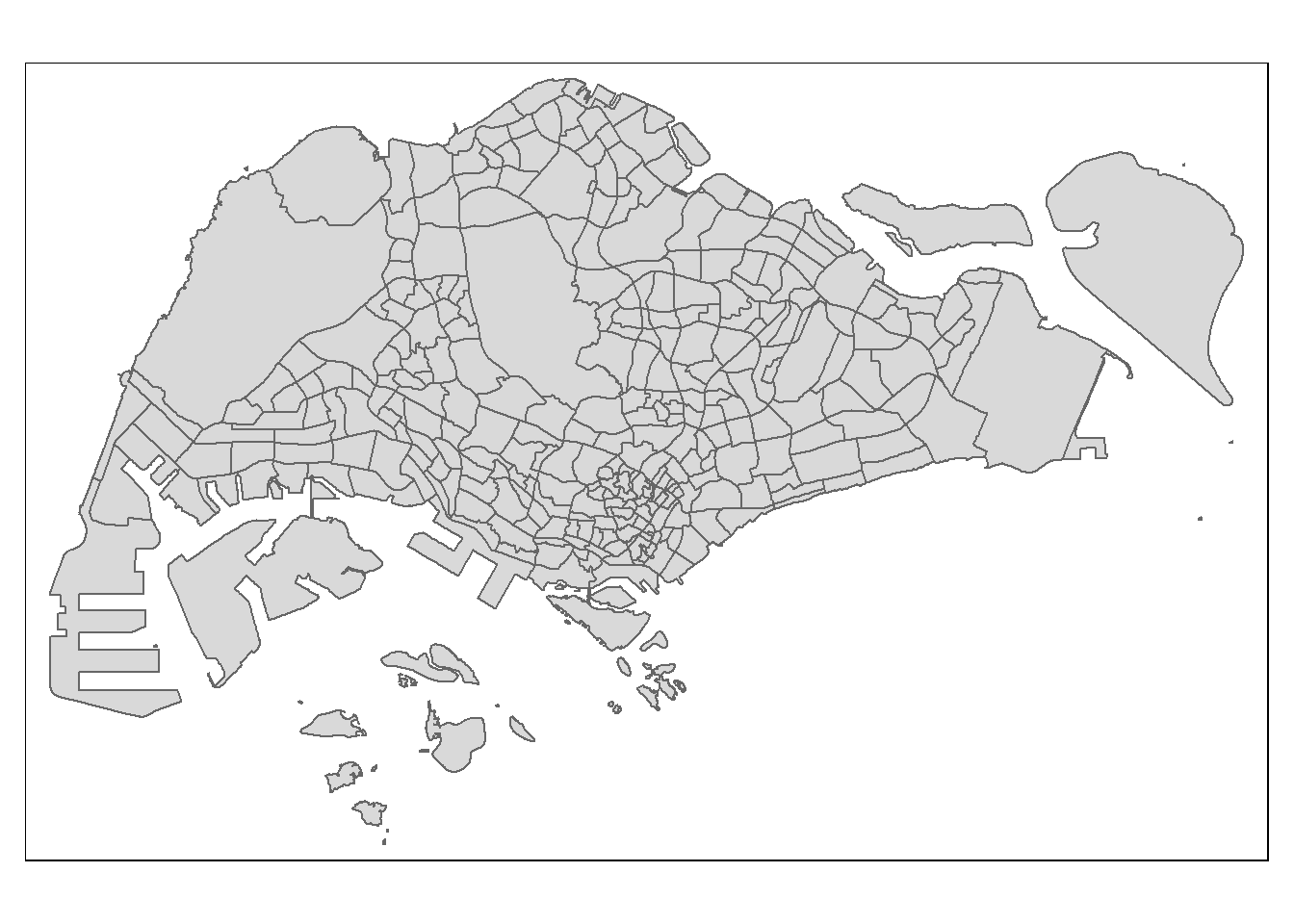
2.2.2.2 Draw a choropleth map using tm_polygons()
To draw a choropleth map showing the geographical distribution of a selected variable by planning subzone, we just need to assign the target variable such as Dependency to tm_polygons().
tm_shape(mpsz_pop2020)+
tm_polygons("DEPENDENCY")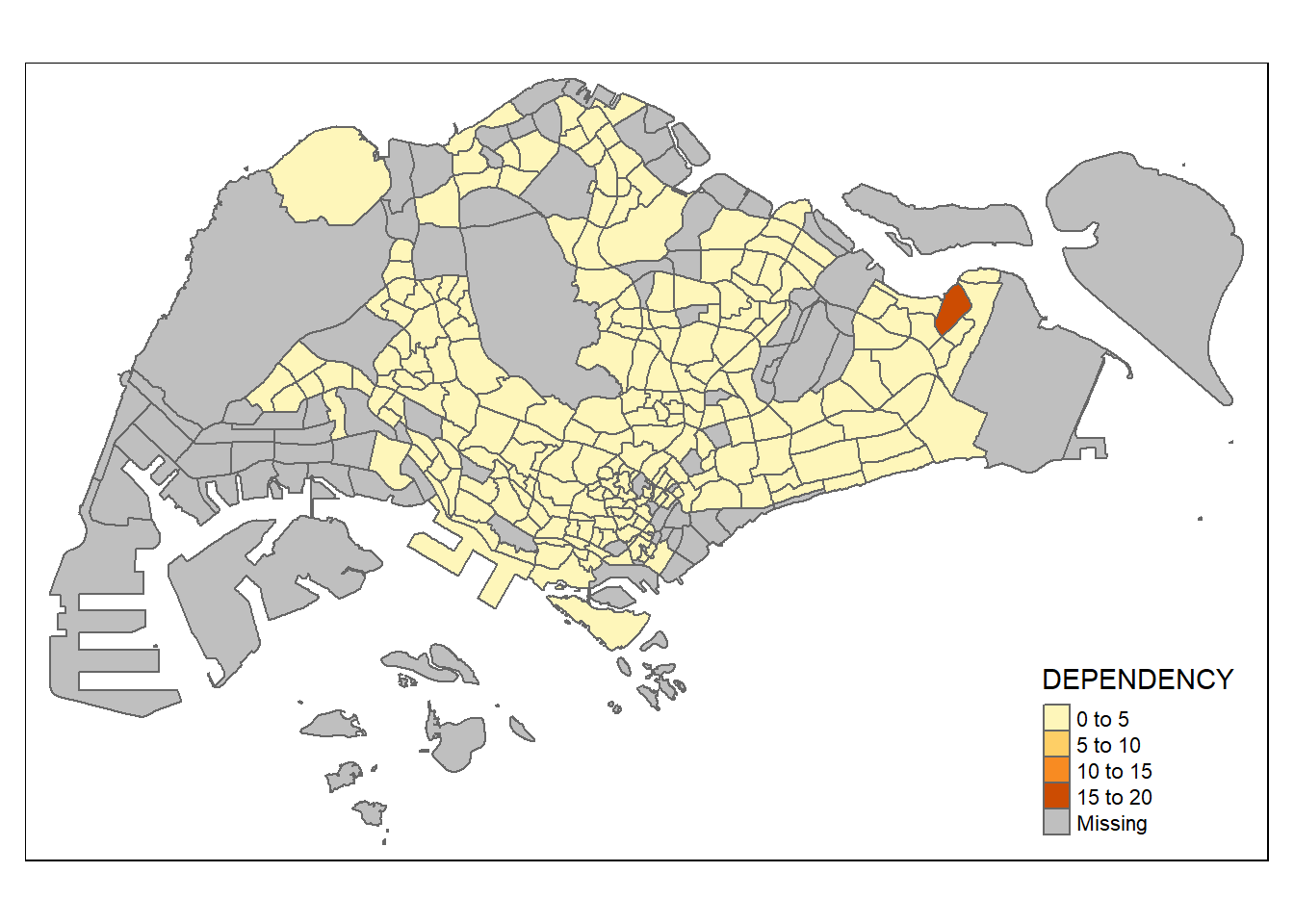
Things to note from tm_polygons():
The default interval binning used to draw the choropleth map is called “pretty”. A detailed discussion of the data classification methods supported by tmap will be provided in sub-section 2.2.3.
The default colour scheme used is
YlOrRdof ColorBrewer. You will learn more about the color scheme in sub-section 2.2.4.By default, Missing value will be shaded in grey.
2.2.2.3 Drawing a choropleth map using tm_fill() and tm_border()
Actually, tm_polygons() is a wraper of tm_fill() and tm_border(). tm_fill() shades the polygons by using the default colour scheme and tm_borders() adds the borders of the shapefile onto the choropleth map.
The code chunk below draws a choropleth map by using tm_fill() alone.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY")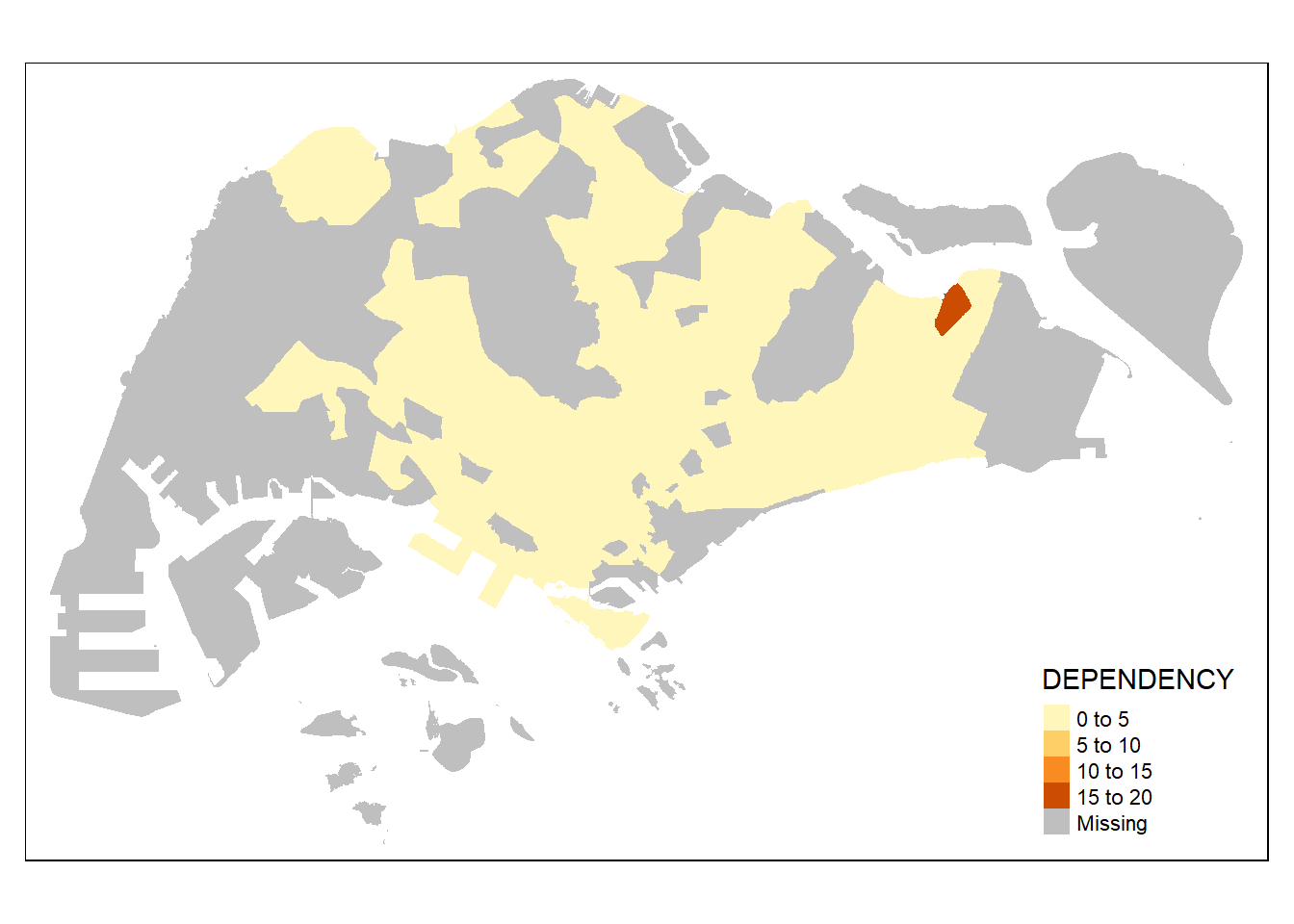
To add the boundary of the planning subzones, tm_borders will be used as shown in the code chunk below.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY") +
tm_borders(lwd = 0.1, alpha = 1)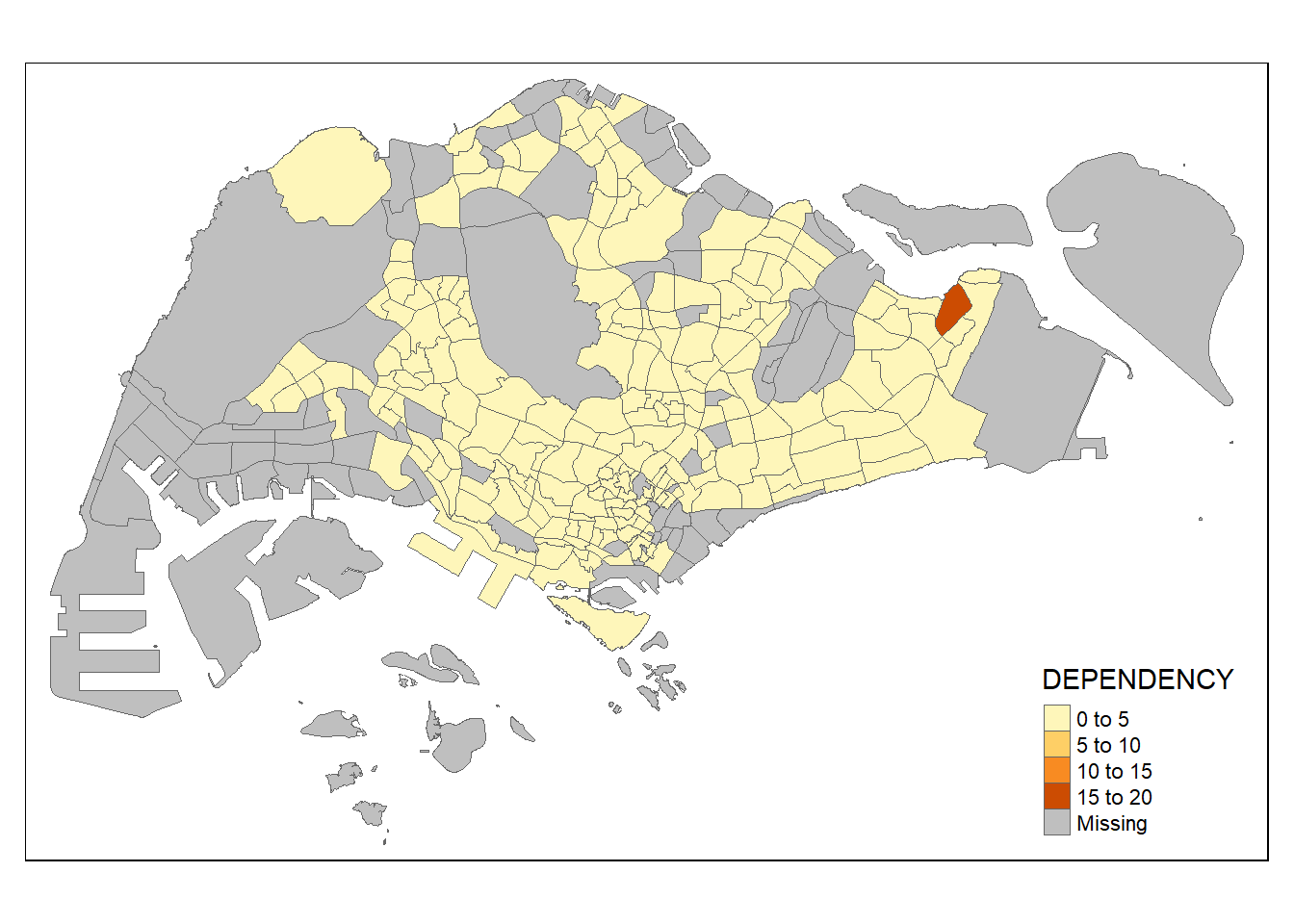
Notice that light-gray border lines have been added on the choropleth map.
The alpha argument is used to define transparency number between 0 (totally transparent) and 1 (obaque). By default, the alpha value of the col is used (normally 1).
Beside alpha argument, there are three other arguments for tm_borders(), they are:
col = border colour,
lwd = border line width. The default is 1, and
lty = border line type. The default is “solid”.
2.2.3 Data classification methods of tmap
Most choropleth maps employ some methods of data classification. The point of classification is to take a large number of observations and group them into data ranges or classes.
tmap provides a total ten data classification methods, namely: fixed, sd, equal, pretty (default), quantile, kmeans, hclust, bclust, fisher, and jenks.
style = pretty: Rounds interval boundaries to whole numbers. Default setting.
style = equal: Splits the variable into intervals of equal length. Should only be used if the variable follows an uniform distribution.
style = quantile: Splits the variable into quantiles. Consequently there are the same number of observations in each interval.
style = jenks: Identifies groups with similar values and maximizes the difference between them.
To define a data classification method, the style argument of tm_fill() or tm_polygons() will be used.
2.2.3.1 Plot choropleth maps with built-in classification methods
The code chunk below shows a quantile data classification that used 5 classes.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
n = 5,
style = "quantile") +
tm_borders(alpha = 0.5)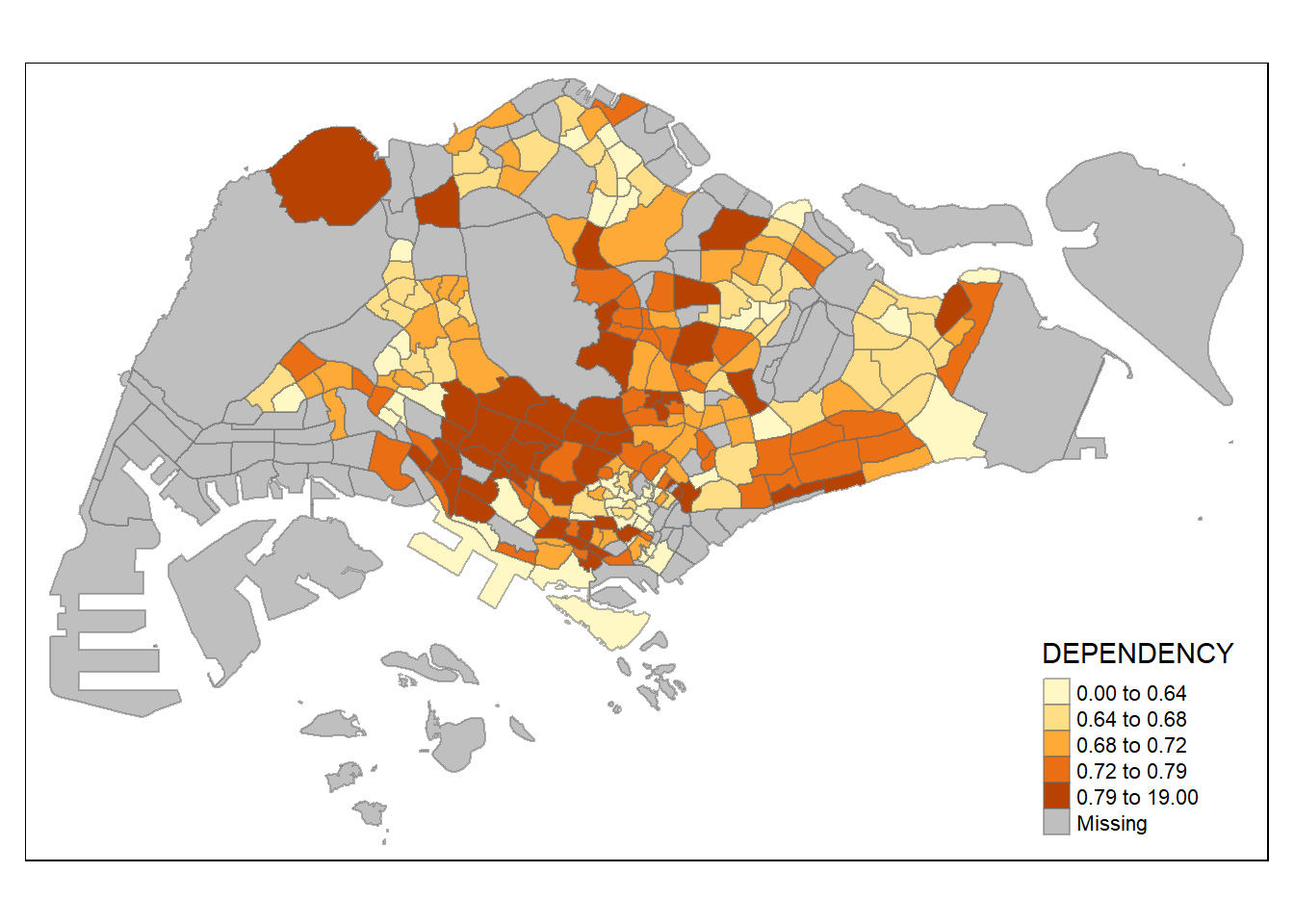
In the code chunk below, equal data classification method is used.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
n = 5,
style = "equal") +
tm_borders(alpha = 0.5)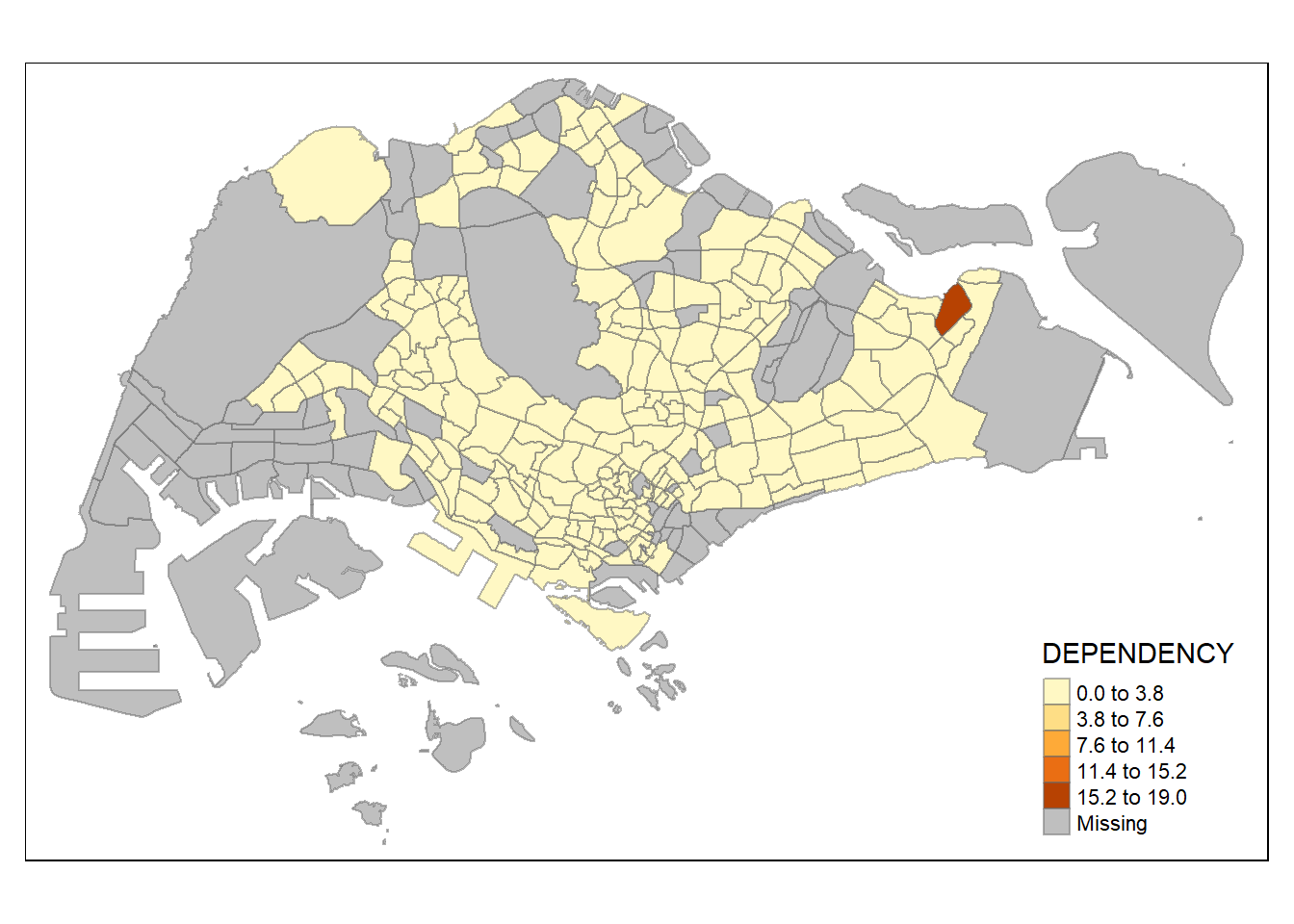
Notice that the distribution of quantile data classification method are more evenly distributed then equal data classification method.
Using pretty style with the following code chunk.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
n = 5,
style = "pretty") +
tm_borders(alpha = 0.5)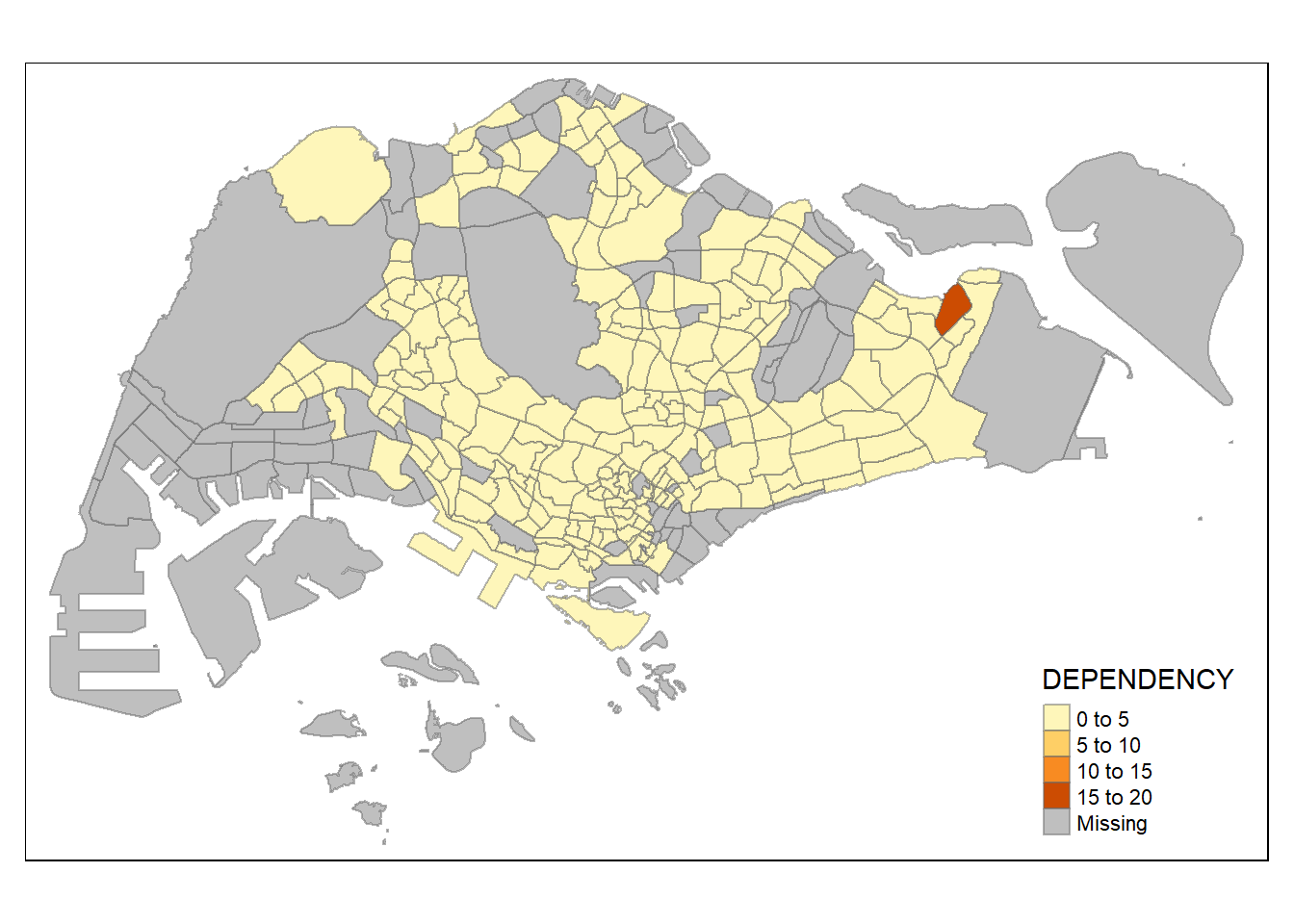
2.2.3.2 Plot choropleth map with customise break
For all the built-in styles, the category breaks are computed internally. In order to override these defaults, the breakpoints can be set explicitly by means of the breaks argument to the tm_fill(). It is important to note that, in tmap the breaks include a minimum and maximum. As a result, in order to end up with n categories, n+1 elements must be specified in the breaks option (the values must be in increasing order).
Before we get started, it is always a good practice to get some descriptive statistics on the variable before setting the break points. Code chunk below will be used to compute and display the descriptive statistics of DEPENDENCY field.
summary(mpsz_pop2020$DEPENDENCY) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.0000 0.6519 0.7025 0.7742 0.7645 19.0000 92 With reference to the results above, we set break point at 0.60, 0.70, 0.80, and 0.90. In addition, we also need to include a minimum and maximum, which we set at 0 and 19. Our breaks vector is thus c(0, 0.60, 0.70, 0.80, 0.90, 19.00)
Now, we will plot the choropleth map by using the code chunk below.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
breaks = c(0, 0.60, 0.70, 0.80, 0.90, 19.00)) +
tm_borders(alpha = 0.5)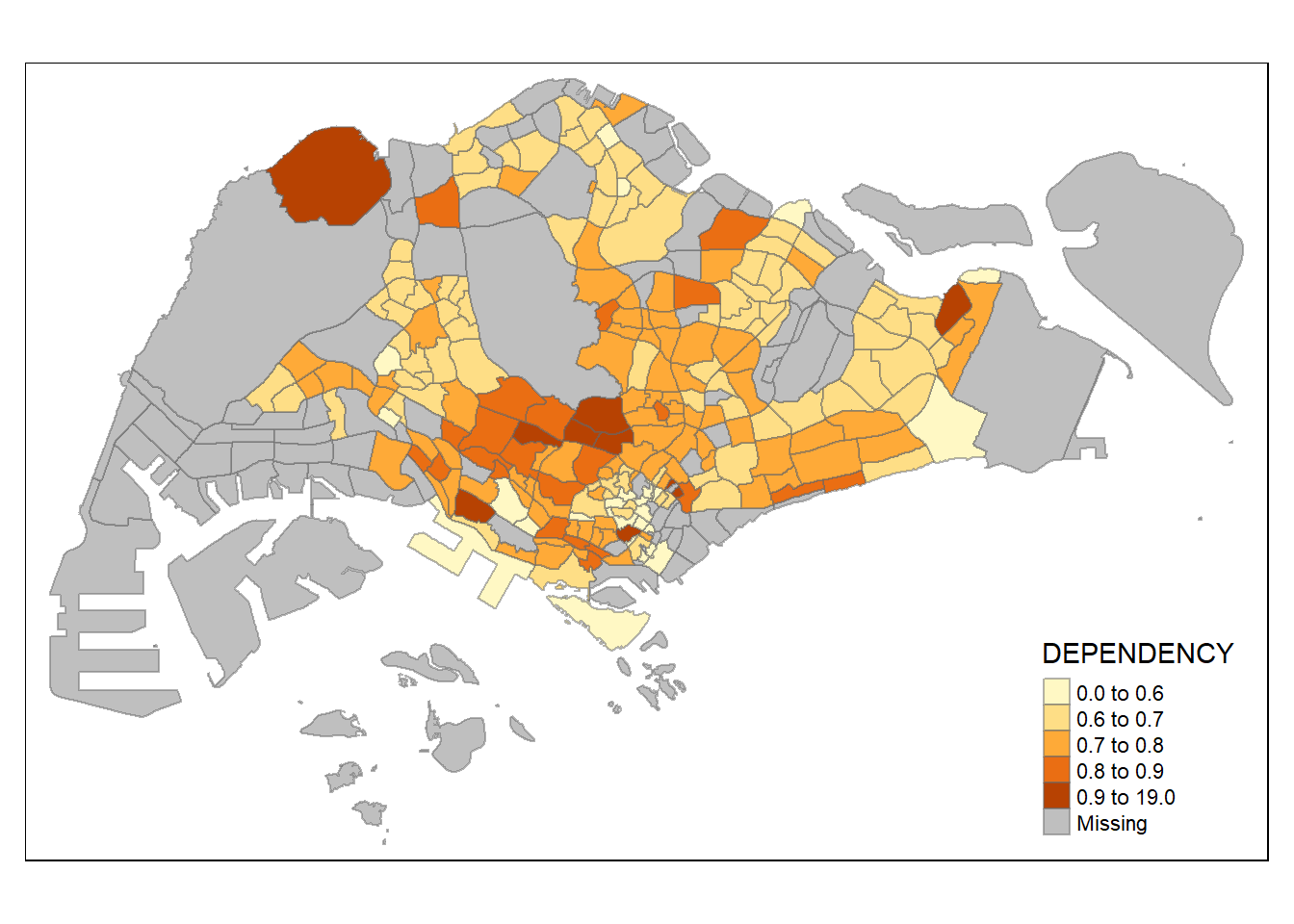
2.2.4 Color Scheme
tmap supports colour ramps either defined by the user or a set of predefined colour ramps from the RColorBrewer package.
2.2.4.1 Use ColourBrewer palette
To change the colour, we assign the preferred colour to palette argument of tm_fill() as shown in the code chunk below.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
n = 6,
style = "quantile",
palette = "Purples") + # Reds, Blues will work as well
tm_borders(alpha = 0.5)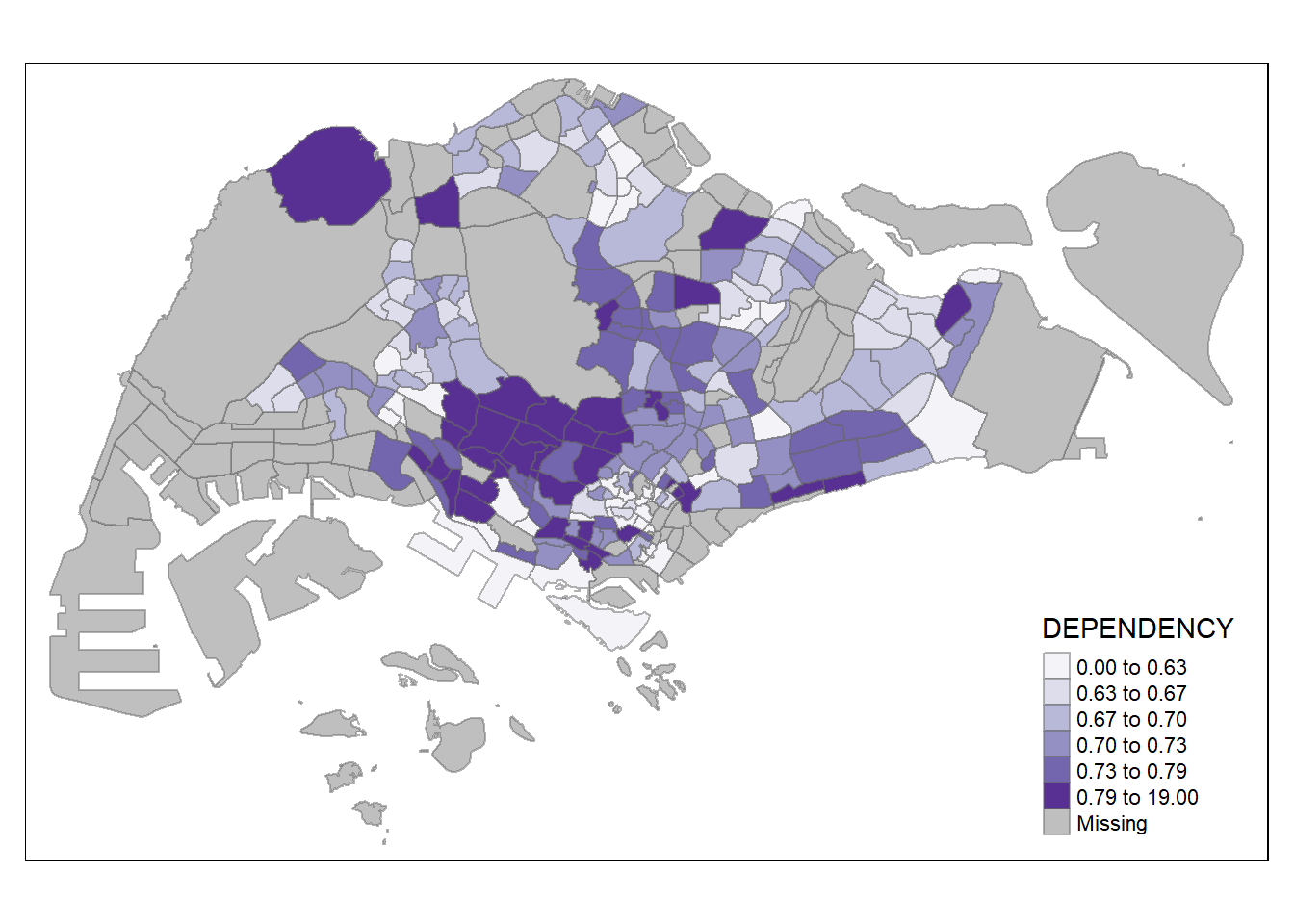
To reverse the colour shading, add a “-” prefix.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
style = "quantile",
palette = "-Greens") +
tm_borders(alpha = 0.5)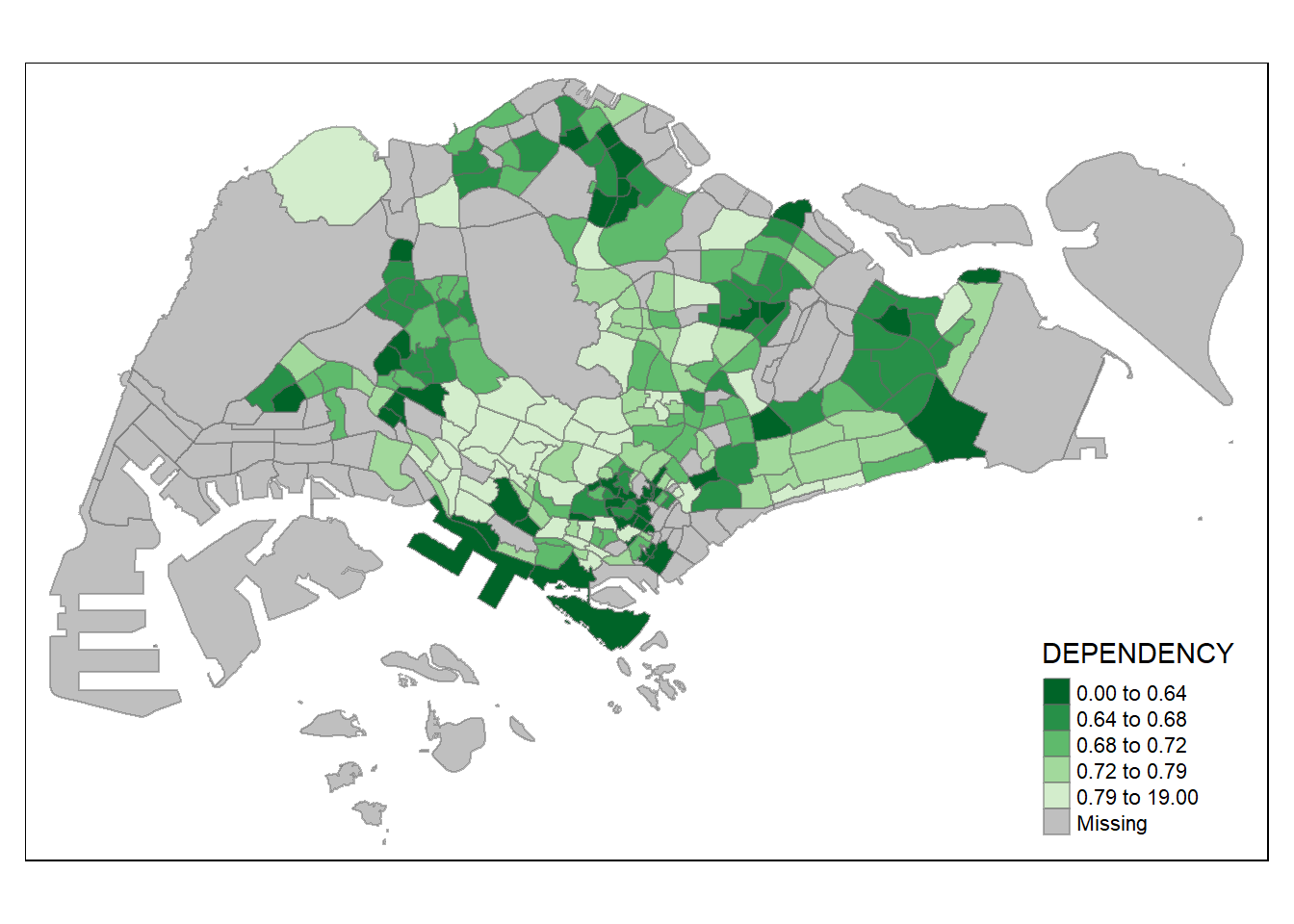
2.2.5 Map Layouts
Map layout refers to the combination of all map elements into a cohensive map. Map elements include among others the objects to be mapped, the title, the scale bar, the compass, margins and aspects ratios. Colour settings and data classification methods covered in the previous section relate to the palette and break-points are used to affect how the map looks.
2.2.5.1 Map Legend
In tmap, several legend options are provided to change the placement, format and appearance of the legend.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
style = "jenks",
palette = "Blues",
legend.hist = TRUE,
legend.is.portrait = TRUE,
legend.hist.z = 0.1) +
tm_layout(main.title = "Distribution of Dependency Ratio by planning subzone \n(Jenks classification)",
main.title.position = "center",
main.title.size = 1,
legend.height = 0.45,
legend.width = 0.35,
legend.outside = FALSE,
legend.position = c("right", "bottom"),
frame = FALSE) +
tm_borders(alpha = 0.5)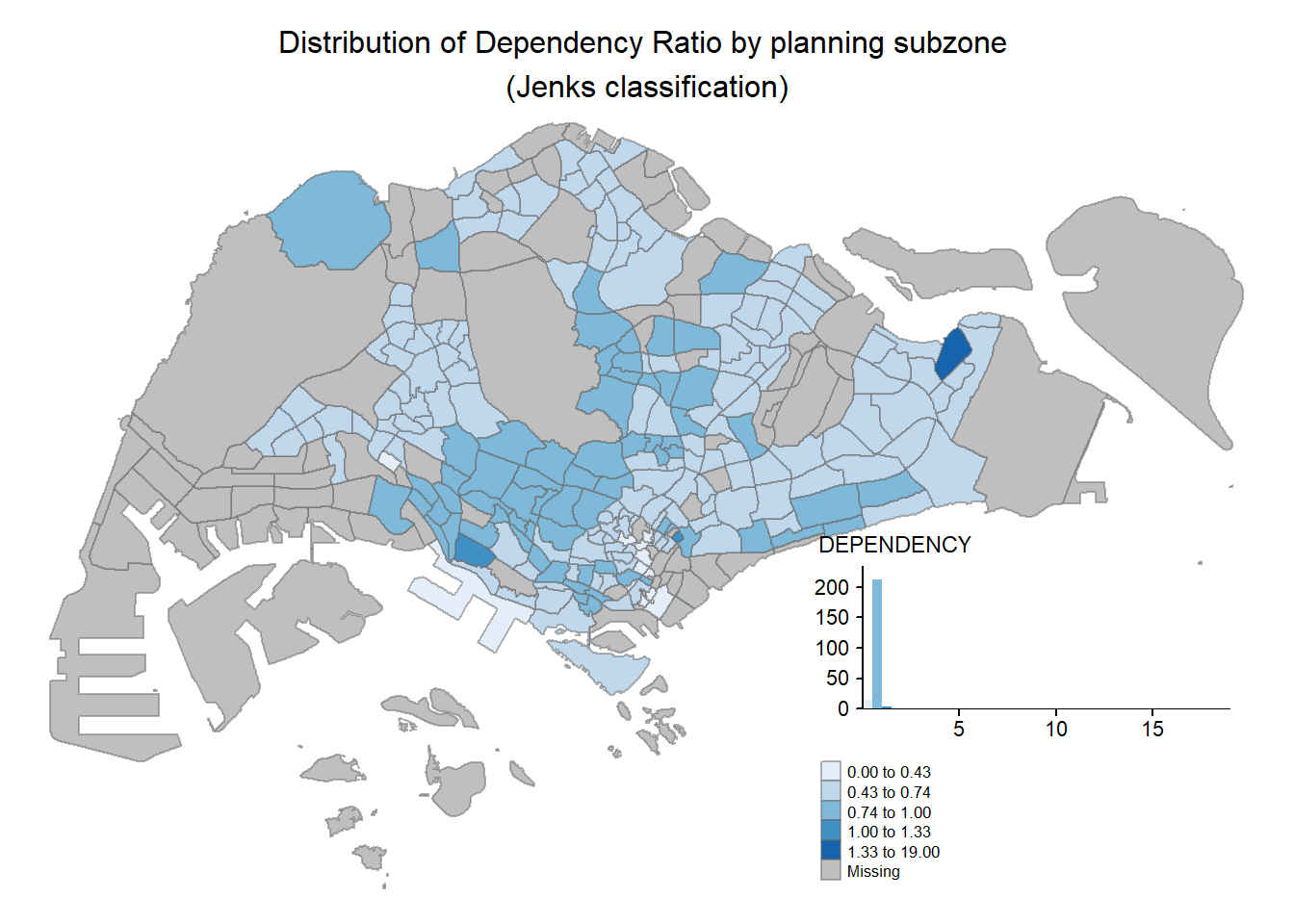
2.2.5.2 Map style
tmap allows a wide variety of layout settings to be changed. They can be called by using tmap_style().
The code chunk below shows the classic style is used.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
style = "quantile",
palette = "-Greens") +
tm_borders(alpha = 0.5) +
tmap_style("classic")tmap style set to "classic"other available styles are: "white", "gray", "natural", "cobalt", "col_blind", "albatross", "beaver", "bw", "watercolor" 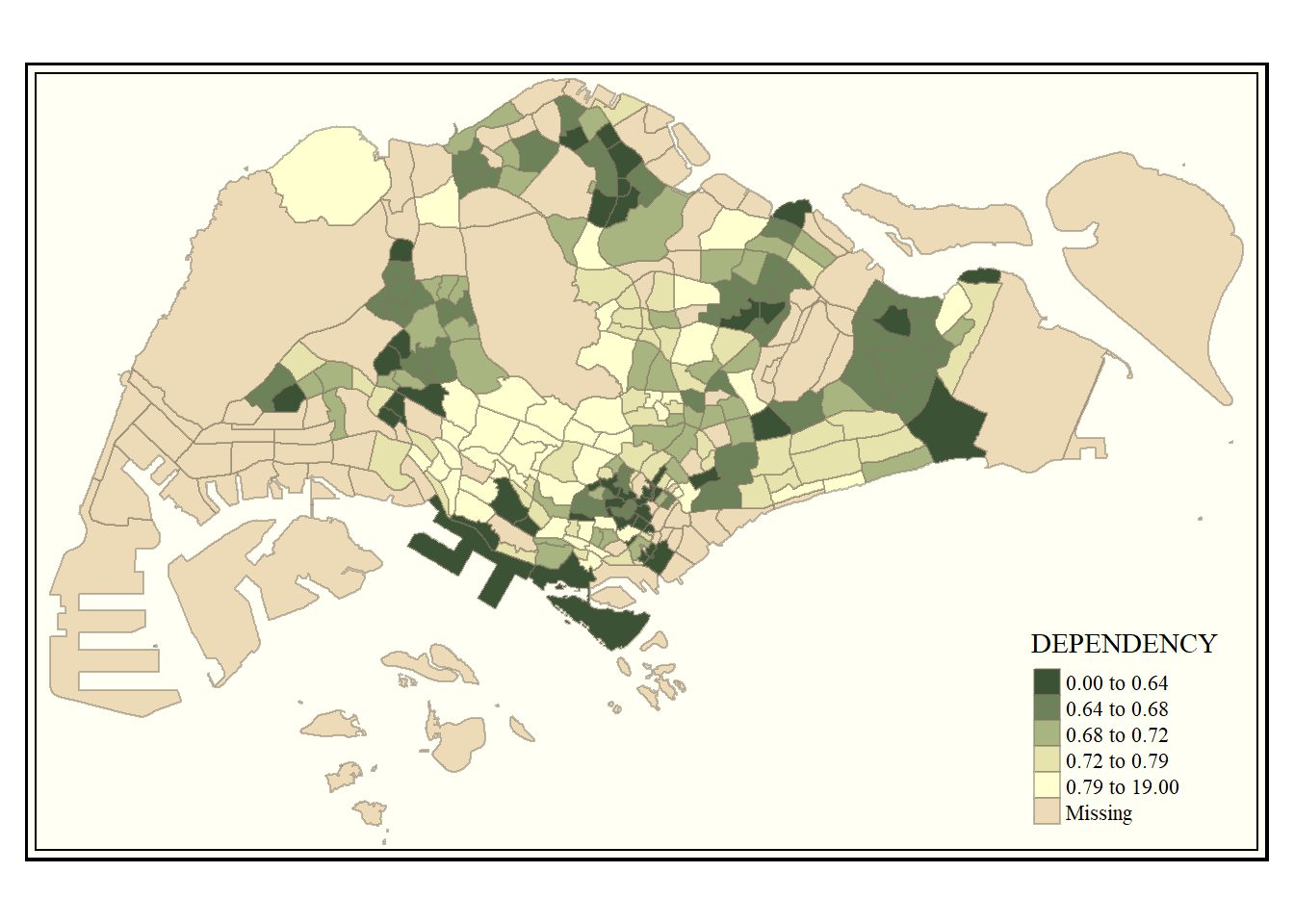
Other available styles are: “white”, “gray”, “natural”, “cobalt”, “col_blind”, “albatross”, “beaver”, “bw”, “watercolor”
2.2.5.3 Cartographic Furniture
tmap also also provides arguments to draw other map furniture such as compass, scale bar and grid lines.
In the code chunk below, tm_compass(), tm_scale_bar() and tm_grid() are used to add compass, scale bar and grid lines onto the choropleth map.
tm_shape(mpsz_pop2020)+
tm_fill("DEPENDENCY",
style = "quantile",
palette = "Blues",
title = "No. of persons") +
tm_layout(main.title = "Distribution of Dependency Ratio \nby planning subzone",
main.title.position = "center",
main.title.size = 1.2,
legend.height = 0.45,
legend.width = 0.35,
frame = TRUE) +
tm_borders(alpha = 0.5) +
tm_compass(type="4star", size = 2) + # can set 8star as well
tm_scale_bar(width = 0.15) +
tm_grid(lwd = 0.1, alpha = 0.2) +
tm_credits("Source: Planning Sub-zone boundary from Urban Redevelopment Authorithy (URA)\n and Population data from Department of Statistics DOS",
position = c("left", "bottom"))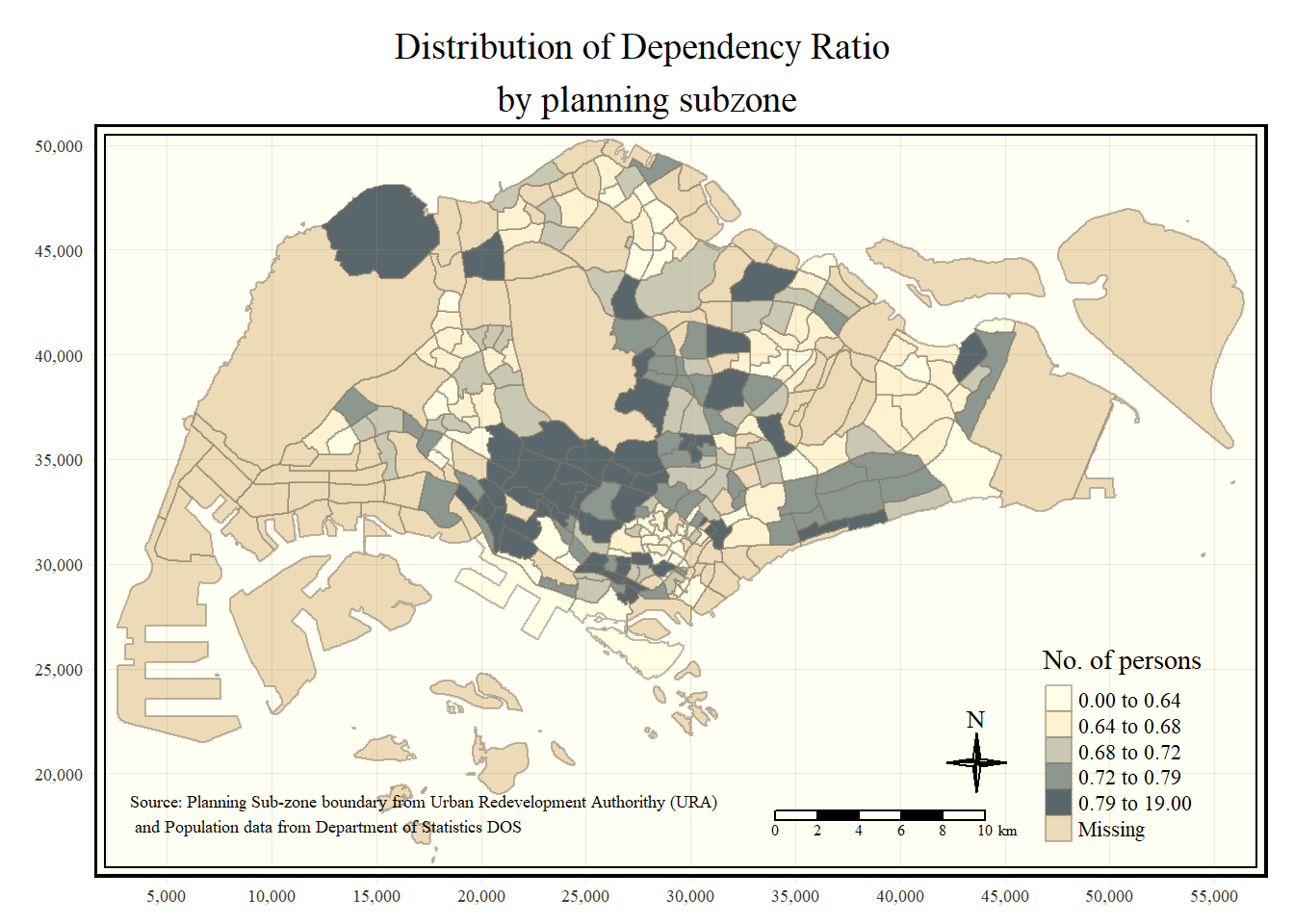
To reset the default style, refer to the code chunk below.
tmap_style("white")tmap style set to "white"other available styles are: "gray", "natural", "cobalt", "col_blind", "albatross", "beaver", "bw", "classic", "watercolor" 2.2.6 Drawing Small Multiple Choropleth Maps
Small multiple maps, also referred to as facet maps, are composed of many maps arrange side-by-side, and sometimes stacked vertically. Small multiple maps enable the visualisation of how spatial relationships change with respect to another variable, such as time.
In tmap, small multiple maps can be plotted in three ways:
by assigning multiple values to at least one of the asthetic arguments,
by defining a group-by variable in tm_facets(), and
by creating multiple stand-alone maps with tmap_arrange().
2.2.6.1 Assign multiple values to at least one of the aesthetic arguments
In this example, small multiple choropleth maps are created by defining ncols in tm_fill()
tm_shape(mpsz_pop2020)+
tm_fill(c("YOUNG", "AGED"),
style = "equal",
palette = "Blues") +
tm_layout(legend.position = c("right", "bottom")) +
tm_borders(alpha = 0.5) +
tmap_style("white")tmap style set to "white"other available styles are: "gray", "natural", "cobalt", "col_blind", "albatross", "beaver", "bw", "classic", "watercolor" 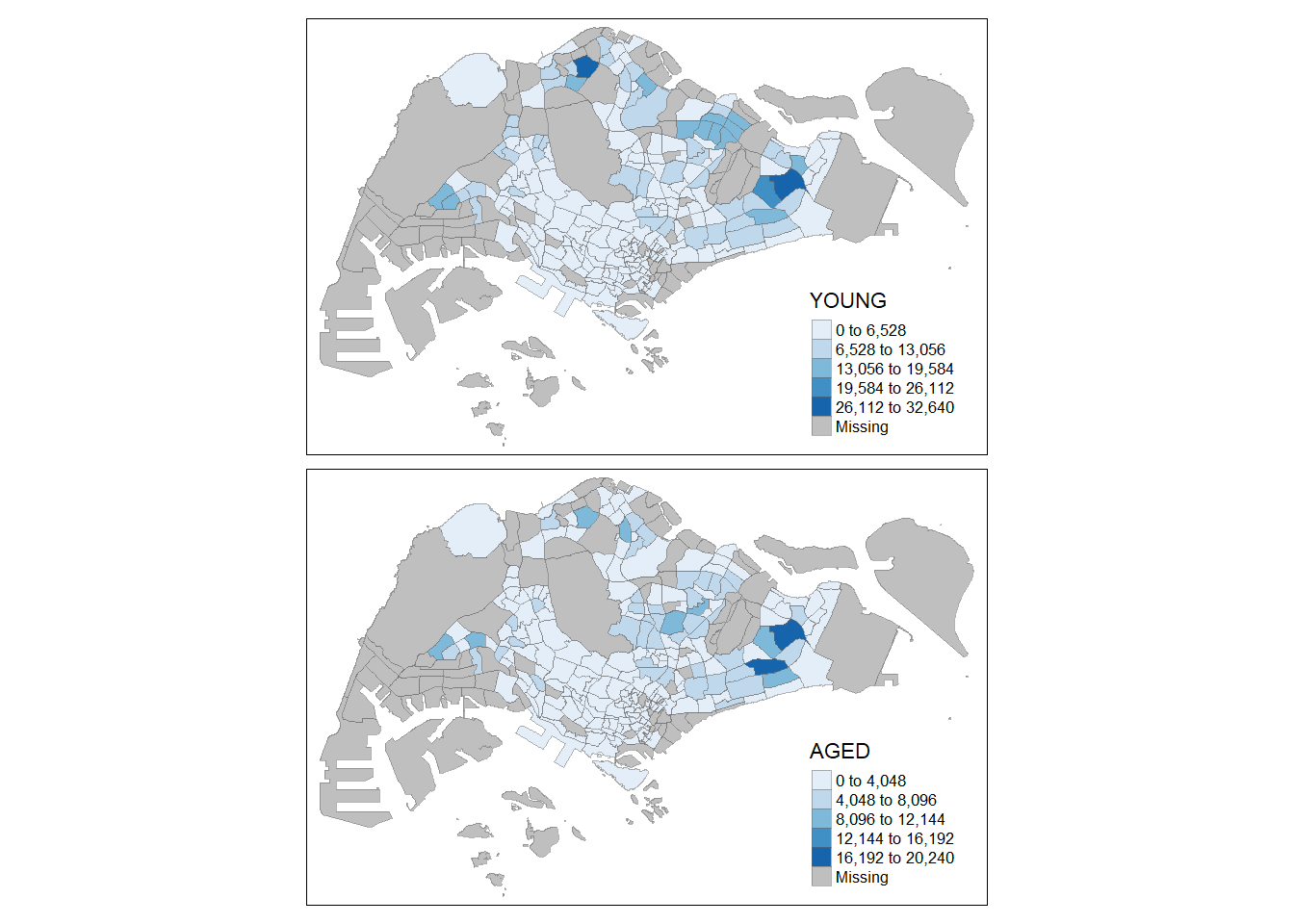
In the following example, small multiple choropleth maps are created by assigning multiple values to at least one of the aesthetic arguments
tm_shape(mpsz_pop2020)+
tm_polygons(c("DEPENDENCY","AGED"),
style = c("equal", "quantile"),
palette = list("Blues","Greens")) +
tm_layout(legend.position = c("right", "bottom"))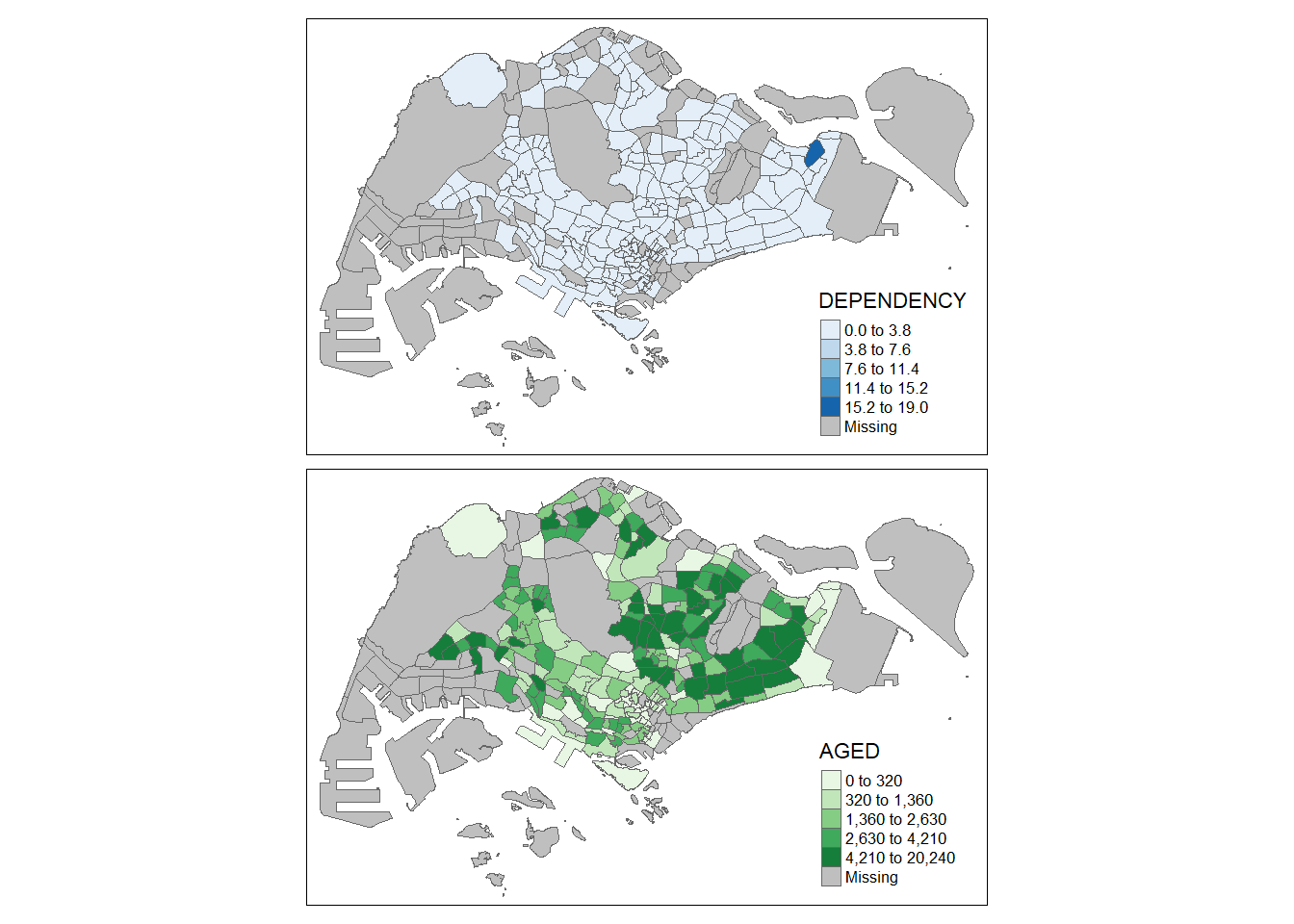
2.2.6.2 Define a group-by variable in tm_facets()
In the following example, multiple small choropleth maps are created by using tm_facets().
tm_shape(mpsz_pop2020) +
tm_fill("DEPENDENCY",
style = "quantile",
palette = "Blues",
thres.poly = 0) +
tm_facets(by="REGION_N",
free.coords=TRUE,
drop.units = TRUE) +
tm_layout(legend.show = FALSE,
title.position = c("center", "center"),
title.size = 20) +
tm_borders(alpha = 0.5)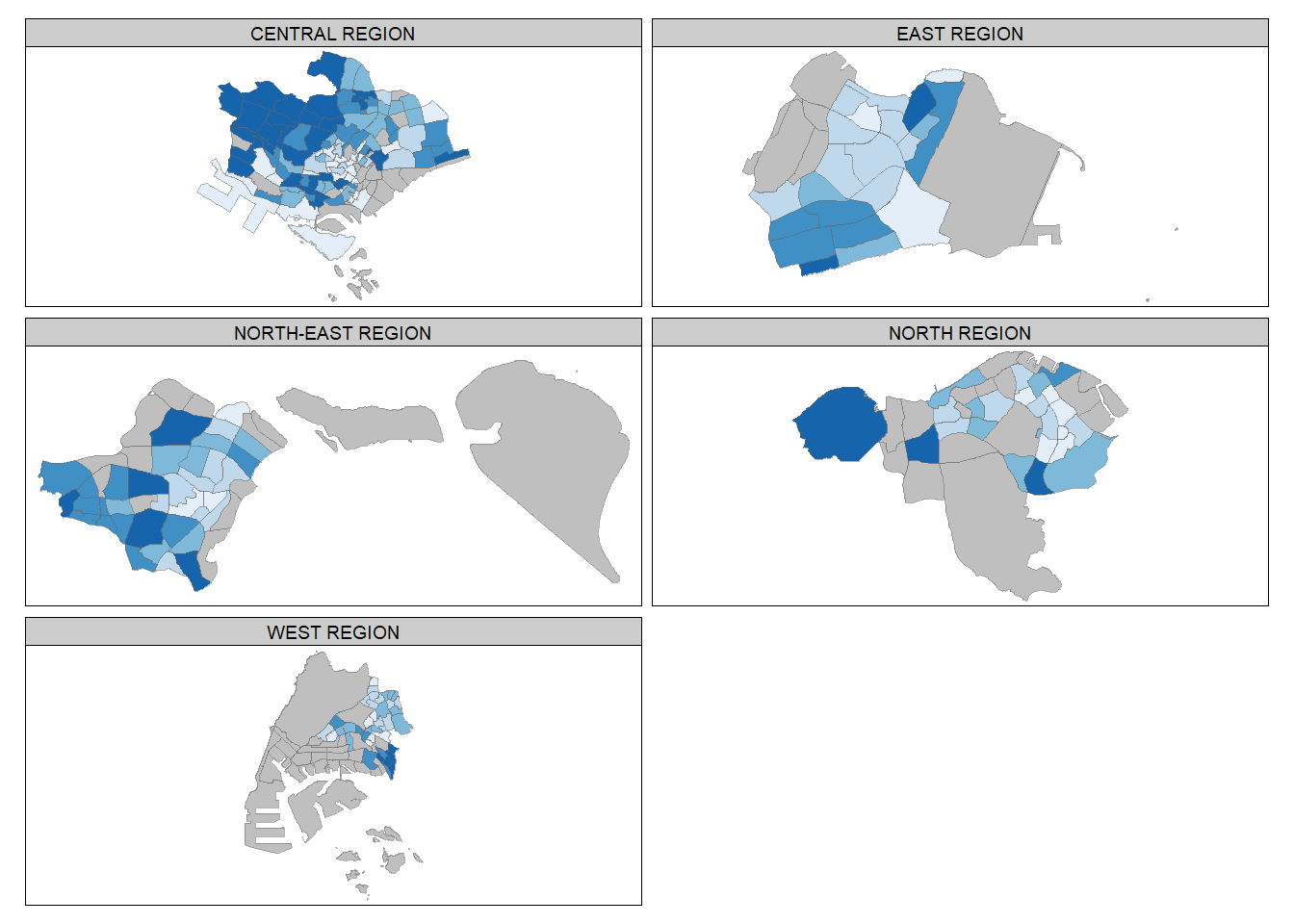
2.2.6.3 Create multiple stand-alone maps with tmap_arrange()
In the following example, multiple small choropleth maps are created by creating multiple stand-alone maps with tmap_arrange().
youngmap <- tm_shape(mpsz_pop2020)+
tm_polygons("YOUNG",
style = "quantile",
palette = "Blues")
agedmap <- tm_shape(mpsz_pop2020)+
tm_polygons("AGED",
style = "quantile",
palette = "Blues")
tmap_arrange(youngmap, agedmap, asp=1, ncol=2)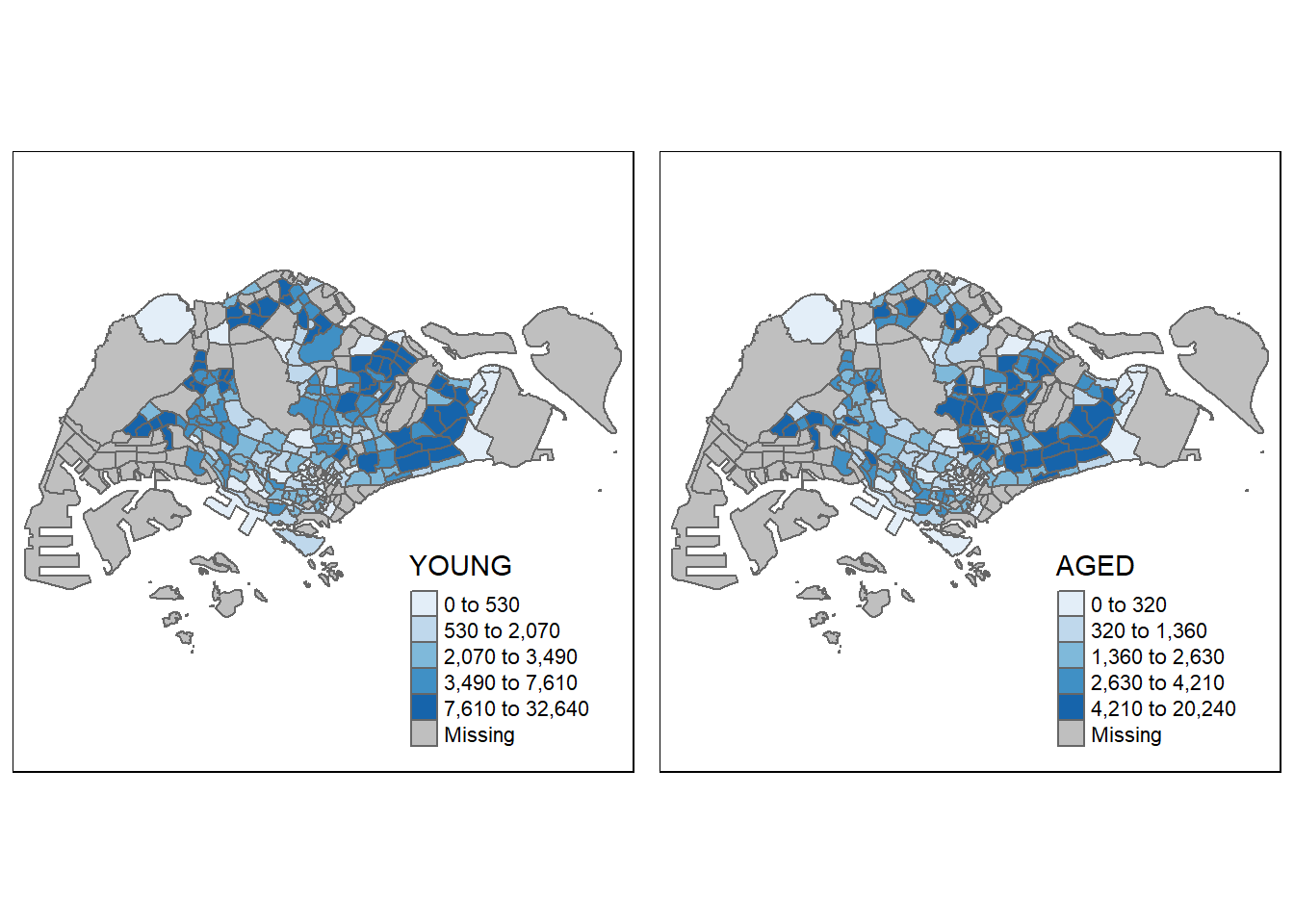
2.2.7 Mapp Spatial Object Meeting a Selection Criterion
Instead of creating small multiple choropleth map, you can also use selection funtion to map spatial objects meeting the selection criterion.
tm_shape(mpsz_pop2020[mpsz_pop2020$REGION_N=="CENTRAL REGION", ])+
tm_fill("DEPENDENCY",
style = "quantile",
palette = "Blues",
legend.hist = TRUE,
legend.is.portrait = TRUE,
legend.hist.z = 0.1) +
tm_layout(legend.outside = TRUE,
legend.height = 0.45,
legend.width = 5.0,
legend.position = c("right", "bottom"),
frame = FALSE) +
tm_borders(alpha = 0.5)Warning in pre_process_gt(x, interactive = interactive, orig_crs =
gm$shape.orig_crs): legend.width controls the width of the legend within a map.
Please use legend.outside.size to control the width of the outside legend